A new technique to build complex custom-designed DNA scaffolds
May 7, 2015
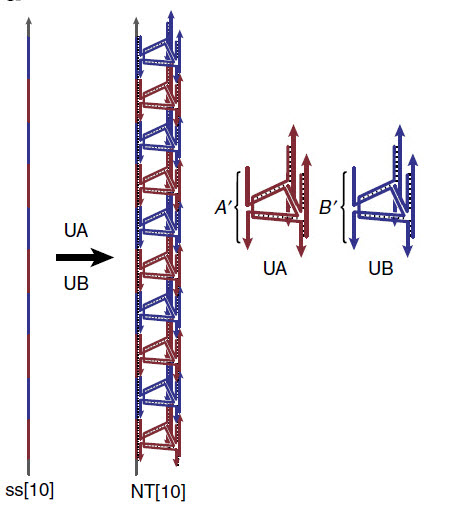
DNA acts as a scaffold for complex nanotube assembly (credit: Graham D. Hamblin et al./Nature Communications)
McGill University researchers have devised a new technique to produce long, custom-designed DNA strands to build nanoscale structures to deliver drugs to targets within the body or take electronic miniaturization to a new level.
Researchers have been assembling and experimenting with DNA structures or “DNA origami” for years, as KurzweilAI has reported. But as these applications continue to develop, they require increasingly large and complex strands of DNA. It can take hundreds of these short strands to assemble nanotubes for applications such as smart drug-delivery systems.
That poses a problem: automated systems used for making synthetic DNA can’t produce strands containing more than about 100 bases (the chemicals that link up to form the DNA strands).
Now in new research published May 5 in Nature Communications, McGill University chemistry professor Hanadi Sleiman and associates report a new technique to create much longer strands of DNA, including custom-designed sequence patterns. This approach also produces large amounts of these longer strands in just a few hours, making the process potentially more economical and commercially viable than existing techniques.
The new method involves piecing together small strands one after the other, so that they attach into a longer DNA strand with the help of an enzyme known as ligase. A second enzyme, polymerase, is then used to generate many copies of the long DNA strand, yielding larger volumes of the material. The polymerase process has the added advantage of correcting any errors that may have been introduced into the sequence, amplifying only the correctly sequenced, full-length product.
Designer DNA materials
The team used these strands as a scaffold to make DNA nanotubes, demonstrating that the technique allows the length and functions of the tubes to be precisely programmed. “In the end, what we get is a long, synthetic DNA strand with exactly the sequence of bases that we want, and with exactly as many repeat units as we want,” explains Sleiman.
“This work opens the door toward a new design strategy in DNA nanotechnology,” she says. “This could provide access to designer DNA materials that are economical and can compete with cheaper but less versatile technologies. In the future, uses could range from customized gene and protein synthesis, to applications in nanoelectronics, nano-optics, and medicine, including diagnosis and therapy.”
Funding for the research was provided by the Natural Sciences and Engineering Research Council of Canada, the Fonds de recherché du Québec-Nature et technologies, the Canada Foundation for Innovation, the Canadian Institutes of Health Research, and the Centre for Self-Assembled Chemical Structures.
McGill University | A better way to build DNA scaffolds
Abstract of Sequential growth of long DNA strands with user-defined patterns for nanostructures and scaffolds
DNA strands of well-defined sequence are valuable in synthetic biology and nanostructure assembly. Drawing inspiration from solid-phase synthesis, here we describe a DNA assembly method that uses time, or order of addition, as a parameter to define structural complexity. DNA building blocks are sequentially added with in-situ ligation, then enzymatic enrichment and isolation. This yields a monodisperse, single-stranded long product (for example, 1,000 bases) with user-defined length and sequence pattern. The building blocks can be repeated with different order of addition, giving different DNA patterns. We organize DNA nanostructures and quantum dots using these backbones. Generally, only a small portion of a DNA structure needs to be addressable, while the rest is purely structural. Scaffolds with specifically placed unique sites in a repeating motif greatly minimize the number of components used, while maintaining addressability. This combination of symmetry and site-specific asymmetry within a DNA strand is easily accomplished with our method.