A super-high-resolution snapshot of RNA folding
November 4, 2016
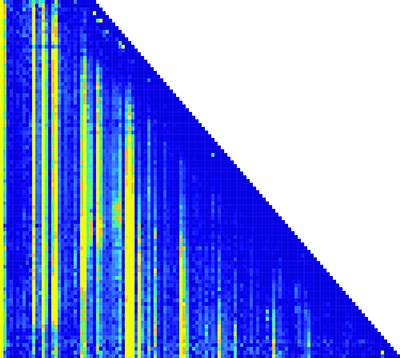
This matrix of the RNA folding pathway shows how the transcription length and nucleotide positions change over time. (Nucleotide position is along the x-axis; transcription length is along the y-axis.) Each pixel in the matrix is a piece of information about the structure of the RNA molecules. (credit: Northwestern University)
Northwestern University engineers have invented a tool to make a super-high-resolution representation of RNA folding as it is being synthesized. It could potentially lead to future discoveries in basic biology, gene expression, RNA viruses, and disease.
Made up of long chains of nucleotides, RNA is responsible for many tasks in the cellular environment, including making proteins, transporting amino acids, gene expression, and carrying messages between DNA and ribosomes. To accomplish all these tasks, RNA folds into complex structures — “one of the biggest, most essential pieces of biology that we know comparatively nothing about,” said Julius B. Lucks, Ph.D, an associate professor of chemical and biological engineering at Northwestern’s McCormick School of Engineering.
RNA folding is an essential requirement to life, but is difficult to investigate because the process occurs rapidly and is extremely hard to measure. Existing technology to image RNA folding is very-low-resolution and can’t image RNA’s individual components rapidly enough to capture these processes.
A molecular microscope
Instead, Lucks’s technology combines two existing components: a next-generation sequencing technique, which is typically used for sequencing human genomes, and a chemistry technique to turn RNA structure measurements into big data.
“Instead of treating it like a genome sequencer, we’re treating it like a molecular microscope to get a massive snapshot,” Lucks said. The technique captures the RNA-folding pathway in a massive dataset. Lucks’s group then uses computational tools to mine and organize the data, which reveals points where the RNA folds and what happens after it folds.
From the structural information that they gather, the researchers can reconstruct a movie of the RNA folding process. The team plans to make the data-analysis component open source, so researchers anywhere can download and run the program.*
Lucks and his team have already used the technology to view the folding of a riboswitch, a segment of RNA that acts as a genetic “light switch” to turn protein expression on or off in response to a molecular signal, in this case fluoride.
Supported by the National Institutes of Health, the research was published online on October 31 in Nature Structural and Molecular Biology.
* The researchers say the technology can be applied to all questions pertaining to RNA, such as how it plays a role in disease. Several notable human diseases, including Ebola, hepatitis C, and measles, are caused by RNA viruses, which are viruses that have RNA as their genetic material. And just as misfolded proteins can cause diseases, such as Alzheimer’s and Parkinson’s, RNA misfolding could also play a role in human illnesses.
Abstract of Cotranscriptional folding of a riboswitch at nucleotide resolution
RNAs can begin to fold immediately as they emerge from RNA polymerase. During cotranscriptional folding, interactions between nascent RNAs and ligands are able to direct the formation of alternative RNA structures, a feature exploited by noncoding RNAs called riboswitches to make gene-regulatory decisions. Despite their importance, cotranscriptional folding pathways have yet to be uncovered with sufficient resolution to reveal how cotranscriptional folding governs RNA structure and function. To access cotranscriptional folding at nucleotide resolution, we extended selective 2′-hydroxyl acylation analyzed by primer-extension sequencing (SHAPE-seq) to measure structural information of nascent RNAs during transcription. Using cotranscriptional SHAPE-seq, we determined how the cotranscriptional folding pathway of the Bacillus cereus crcB fluoride riboswitch undergoes a ligand-dependent bifurcation that delays or promotes terminator formation via a series of coordinated structural transitions. Our results directly link cotranscriptional RNA folding to a genetic decision and establish a framework for cotranscriptional analysis of RNA structure at nucleotide resolution.