An ‘in silico’ method of predicting effectiveness of cognitive enhancers
February 17, 2015
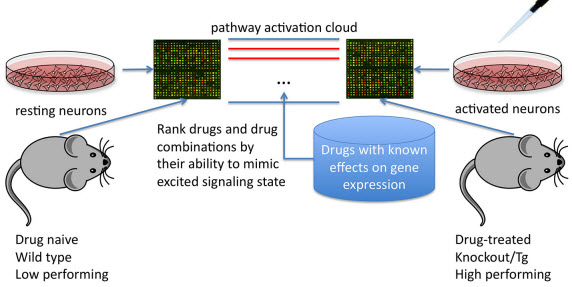
Using differential signaling pathway activation profiles between cell/tissue culture or mice treated and untreated with known cognitive modulators or genetic models of cognitive enhancement to screen for potential nootropic compounds. (credit: Leslie C. Jellen et al./Frontiers in Systems Neuroscience)
The Biogerontology Research Foundation (BGRF) has used gene expression data to evaluate activated or suppressed signaling pathways in tissues or neurons of the mouse brain that has been cognitively enhanced with nootropic drugs.
Currently used cognitive enhancers are those that are widely available, rather than optimal for the user, the researchers note. These include drugs typically prescribed for treatment of ADHD (e.g., methylphenidate) and sleep disturbances such as narcolepsy (modafinil). The researchers want to quantify the effects with objective measures and thus predict the efficacy of the many drugs that may enhance various aspects of cognition — before costly preclinical studies and clinical trials are undertaken.
The research, published in open-access Frontiers in Systems Neuroscience, uses an algorithm that maps expression data onto signaling pathways. The collective pathways and their activation form a “signaling pathway cloud” — a biological fingerprint of cognitive enhancement. Drugs can then be screened and ranked based on their ability to minimize, mimic, or exaggerate pathway activation or suppression within that cloud.
“Our current work in predicting the efficacy of drugs and drug combinations in treating and preventing some of the most age-related diseases, suggests that some likely geroprotectors may also enhance cognitive function, said Alex Zhavoronkov, PhD, director of the BGRF. “We are actively seeking academic and industry collaborators for this exciting neuroscience project.”
The BGRF is a UK-based charity committed to the support of aging research.
Abstract of Screening and personalizing nootropic drugs and cognitive modulator regimens in silico
The go-to cognitive enhancers of today are those that are widely available rather than optimal for the user, including drugs typically prescribed for treatment of ADHD (e.g., methylphenidate) and sleep disturbances such as narcolepsy (modafinil). While highly effective in their intended therapeutic role, performance gains in healthy populations are modest at best and profoundly inconsistent across subgroups and individuals. We propose a method for in silico screening of possible novel cognitive enhancers followed by high-throughput in vivo and in vitro validation. The proposed method uses gene expression data to evaluate the the collection of activated or suppressed signaling pathways in tissues or neurons of the cognitively enhanced brain. An algorithm maps expression data onto signaling pathways and quantifies their individual activation strength. The collective pathways and their activation form what we term the signaling pathway cloud, a biological fingerprint of cognitive enhancement (or any other condition of interest). Drugs can then be screened and ranked based on their ability to minimize, mimic, or exaggerate pathway activation or suppression within that cloud. Using this approach, one may predict the efficacy of many drugs that may enhance various aspects of cognition before costly preclinical studies and clinical trials are undertaken.