As the worm turns: research tracks how an embryo’s brain is assembled
December 7, 2015
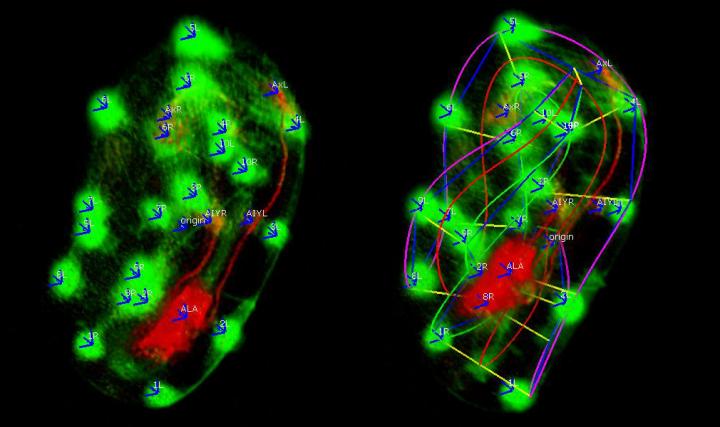
The image on the left shows skin cells (green dots) and neurons (red cell) marking the shape of the embryo. The image on the right shows the skin cells connected by the software to make a computerized model of how the embryo folds and twists. (credit: Hari Shroff, National Institute of Biomedical Imaging and Bioengineering)
New open-source software that can help track the embryonic development and movement of neuronal cells throughout the body of a worm is now available to scientists. The software is described in a paper published in the open access journal, eLife on December 3rd by a research team*.
One significant challenge is determining the formation of complex neuronal structures made up of billions of cells in the human brain. As with many biological challenges, researchers are first examining this question in simpler organisms, such as worms.
Although scientists have identified a number of important proteins that determine how neurons navigate during brain formation, it’s largely unknown how all of these proteins interact in a living organism.
Model animals, despite their differences from humans, have already revealed much about human physiology because they are much simpler and easier to understand. In this case, researchers chose Caenorhabditis elegans (C. elegans), because it has only 302 neurons, 222 of which form while the worm is still an embryo.
While some of these neurons go to the worm nerve ring (brain) they also spread along the ventral nerve cord, which is broadly analogous to the spinal cord in humans. The worm even has its own versions of many of the same proteins used to direct brain formation in more complex organisms such as flies, mice, or humans.
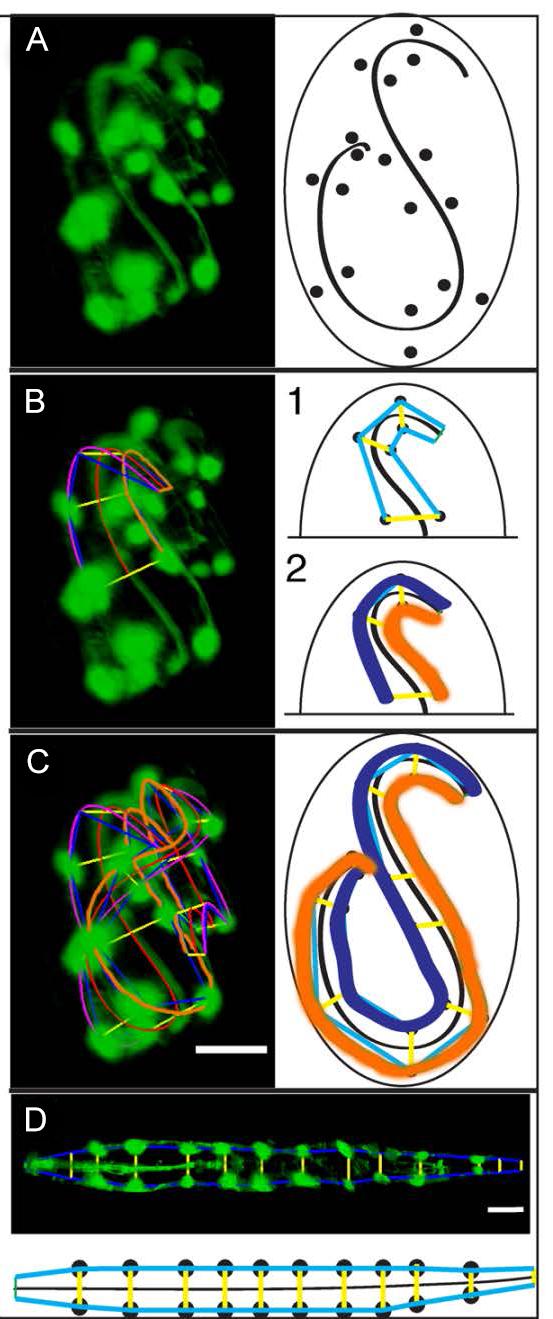
These four images show the step by step process the computer program goes through to the “untwist” a worm image. First, (Image D) the computer identifies the cells to track. Then (Image E) the computer begins to create a lattice that traces the shape of the worm. Once the computer has traced the lattice, it can create a 3D model of the worm embryo (Image F). Finally, it can untwist the model (Image G). (credit: Hari Shroff, National Institute of Biomedical Imaging and Bioengineering)
However, following neurons as they travel through the worm during its embryonic development is not as simple as it might seem. The first challenge was to create new microscopes that could record the embryogenesis of these worms without damaging them through too much light exposure while still getting the resolution needed to clearly see individual cells.
Shroff and his team at NIBIB, in collaboration with Daniel Colon-Ramos at Yale University and Zhirong Bao at Sloan-Kettering, tackled this problem by developing new microscopes that improved the speed and resolution at which they could image worm embryonic development. (Read more)
The second problem was that during development, the worm begins to “twitch,” moving around inside the egg. The folding and twisting makes it hard to track cells and parse out movement. For example, if a neuron moves in the span of a couple of minutes, is it because the embryo twisted or because the neuron actually changed position within the embryo?
Understanding the mechanisms that move neurons to their final destination is an important factor in understanding how brains form–and is difficult to determine without knowing where and how a neuron is moving. Finally, it can be challenging to determine where a neuron is in 3D space while looking at a two-dimensional image–especially of a worm that’s folded up.
The worm embryo is normally transparent, but the researchers made several cells in the embryo glow with fluorescent proteins to act as markers.
When a microscopic image of these cells is fed into the program, the computer identifies each cell and uses the information to create a model of the worm, which it then computationally “untwists” to generate a straightened image. The program also enables a user to check the accuracy of the computer model and edit it when any mistakes are discovered.
“In addition, users can also mark cells or structures within the worm embryo they want the program to track, allowing the users to follow the position of a cell as it moves and grows in the developing embryo. This feature could help scientists understand how certain cells develop into neurons, as opposed to other types of cells, and what factors influence the development of the brain and neuronal structure.
A worm atlas
Shroff and his colleagues say that such technology will be pivotal in their project to create a 4D neurodevelopmental “worm atlas,” (see also http://www.wormguides.org) that attempts to catalog the formation of the worm nervous system.
This catalog will be the first comprehensive view of how an entire nervous system develops, and the researchers believe that it will be helpful in understanding the fundamental mechanisms by which all nervous systems, including ours, assemble. They also expect that some of the concepts developed, such as the approach taken to combine neuronal data from multiple embryos, can be applied to additional model organisms besides the worm.
* Researchers at the National Institute of Biomedical Imaging and Bioengineering (NIBIB) and the Center for Information Technology (CIT); along with Memorial Sloan-Kettering Institute, New York City; Yale University, New Haven, Connecticut; Zhejiang University, China; and the University of Connecticut Health Center, Farmington. NIBIB is part of the National Institutes of Health.
NIBIB | C. Elegans Embryo Development
Abstract of Untwisting the Caenorhabditis elegans embryo
The nematode Caenorhabditis elegans possesses a simple embryonic nervous system comprising 222 neurons, a number small enough that the growth of each cell could be followed to provide a systems-level view of development. However, studies of single cell development have largely been conducted in fixed or pre-twitching live embryos, because of technical difficulties associated with embryo movement in late embryogenesis. We present open source untwisting and annotation software which allows the investigation of neurodevelopmental events in post-twitching embryos, and apply them to track the 3D positions of seam cells, neurons, and neurites in multiple elongating embryos. The detailed positional information we obtained enabled us to develop a composite model showing movement of these cells and neurites in an “average” worm embryo. The untwisting and cell tracking capability we demonstrate provides a foundation on which to catalog C. elegans neurodevelopment, allowing interrogation of developmental events in previously inaccessible periods of embryogenesis.