Bacteria are social microorganisms: MIT researchers
September 12, 2012
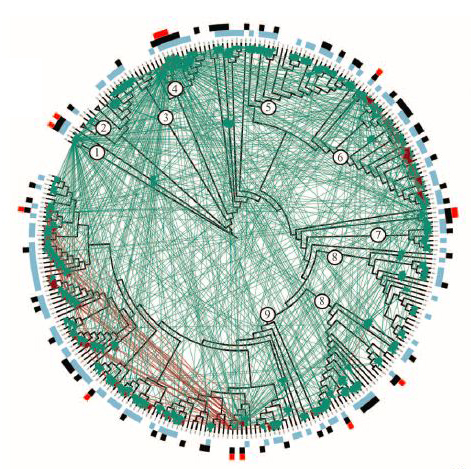
Distribution of antagonistic interactions in relationship to Vibrio phylogeny and genetic distance. (A) Phylogeny of Vibrio isolates based on six housekeeping genes, with outer, colored rings highlighting antagonists and sensitive strains. Green arrows connect antagonists to sensitive strains. Circles identify the most recent common ancestor (MRCA) of previously identified ecologically cohesive populations: 1, V. ordalii; 2, V. fischeri; 3, V. breoganii; 4, V. alginolyticus; 5, V. sp. F12; 6, V. crassostreae; 7, V. cyclotrophicus; 8, V. tasmaniensis; 9, V. splendidus. (Credit: Otto X. Cordero et al./Science)
Bacteria can have social structures similar to plants and animals, new MIT research reveals.
Bacteria can produce chemical compounds that kill or slow the growth of other populations of bacteria in the environment, but not harm their own.
“Bacteria typically have been considered purely selfish organisms and bacterial populations as groups of clones,” said Otto Cordero, a theoretical biologist and lead researcher on the paper.
“This result contrasts with what we know about animal and plant populations, in which individuals can divide labors, perform different complementary roles and act synergistically.”
Cordero and colleagues from MIT, along with researchers from the French Research Institute for Exploitation of the Sea and Woods Hole Oceanographic Institution in Massachusetts, studied whether population-level organization exists for bacteria in the wild.
They reasoned social structure can reduce conflict within populations of plants and animals and determine aggression towards competing biological populations. “Think of a population of lions in the Serengeti or a population of fish in a lake,” said Cordero. But could the same be true for populations of bacteria?
Chemical warfare
The researchers found evidence of use of antibiotics by looking at direct, aggressive competition between ecological populations of bacteria. They reconstructed a large network of bacterial fights — or antibiotic-mediated interactions — between bacteria from the ocean.
The scientists analyzed interactions called interference competitions, wherein bacteria produce antibiotics as a means of chemical warfare, to gain a competitive edge by directly hindering the survival of potential competitors.
This typically occurs when bacteria compete for the same portion of habitat.
The researchers assembled an all-against-all battleground for 185 closely-related, but distinct, members of an ocean-based family of bacteria called Vibrionaceae. They measured bacterial compounds produced by Vibrio isolates that directly antagonized other Vibrio isolates.
The framework provided Cordero and colleagues an opportunity to examine about 35,000 possible antibiotic-mediated interactions.
Socially cohesive units
The researchers found that ecologically delineated bacterial populations act as socially cohesive units. “In these populations, a few individuals produced antibiotics to which closely related individuals in the population were resistant, whereas individuals in other populations were sensitive,” said Cordero.
“Those individuals that don’t produce antibiotics can benefit from association with the producers, because they are resistant,” added Cordero. “In other words, antibiotics have a social effect, because they can benefit the population as a whole.”
The findings may help scientists answer questions about the natural role of antibiotics in human contexts.
“The research has the potential to bridge gaps in our understanding of the relationships between plants and humans and their non-disease- and disease-causing bacterial flora,” said Robert Fleischmann, a program director in the Division of Biological Infrastructure for the National Science Foundation.
“We use antibiotics to kill pathogenic microbes, which cause harm to humans and animals,” said Polz. “As an unfortunate side effect, this has lead to the widespread buildup of resistance, particularly in hospitals where pathogens and humans encounter each other often.”
In addition, the results help scientists make sense of why closely related bacteria are so diverse in their gene content. Part of the answer, they say, is that the diversity allows the bacteria to play different social roles.
Social differentiation, for example, could mitigate the negative effects of two species competing for the same limiting resource — food or habitat, for instance — and generate population level behavior that emerges from the interaction between close relatives.
“Microbiology builds on the study of pure cultures,” said Cordero, “that is genotypes isolated from their population. Our work shows that we need to start focusing on population based phenomena to better understand what these organisms are doing in the wild.”
The National Science Foundation funded the research.