A molecular ‘switch’ to reprogram control pathways in cells
August 20, 2013
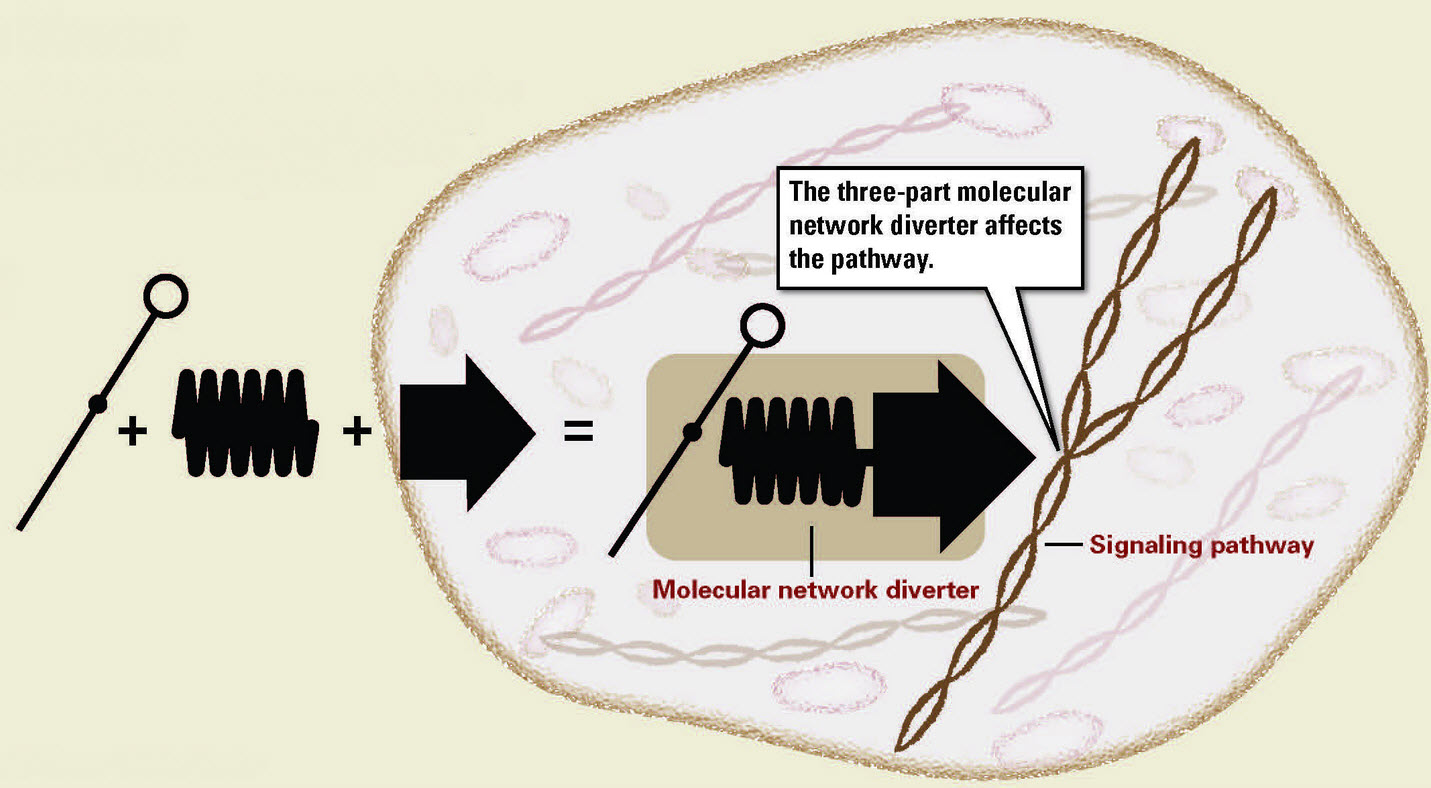
A modular technology that uses three biological components to control a signaling pathway inside a cell: the transducer (left), an RNA-based system that gathers information about the chemical environment of the cell; the promoter (spring), the molecular agent that helps to initiate and modulate the desired change; and the pathway regulator (arrow), which finds the appropriate point in the cell’s signaling pathway to make the intervention (credit: Rick Nobles/Stanford Engineering)
A Stanford University bioengineer has helped develop a technology dubbed a “molecular network diverter” that can tweak the control systems that regulate the inner workings of cells, pointing the way toward future medical interventions that could switch off diseased states or turn on healthy processes.
This molecular diverter uses the concerted action of three biological sub-systems to redirect signaling pathways — complex networks of molecular interactions that orchestrate the cellular machinery.
Controlling signaling pathways that cancer, other diseases
The experiments described by Christina Smolke, PhD, associate professor of bioengineering, and associates were performed on yeast cells. But the principles and practices embodied in the molecular network diverter apply to signaling pathways that control the development, reproduction and death of all cells. When these signaling pathways go awry in humans, for instance, such malfunctions can cause many types of cancer as well as other diseases.
“We’re doing this in yeast, but there’s a lot of conservation, or similarity, of these pathways in higher organisms,” Smolke said. “The next step, now that we’ve shown this in simpler systems, is to take this technology into human cell cultures.”
The team’s initial goal was to control the mating behavior of yeast, an activity that, in nature, is influenced by the presence or absence of pheromones, which are naturally occurring odorless substances that can trigger responses from the opposite sex.
Engineering cell behavior
In a series of experiments, Smolke and her collaborators tried various techniques to induce or inhibit yeast mating behavior irrespective of pheromone activity.
At first they found that the various techniques they used canceled each other out. But through computational modeling and fine-tuning of the chemical components they were able to build the molecular network diverter by joining three techniques into a unified technology whose elements they call:
- The transducer, an RNA-based system that gathers information about the chemical environment of the cell.
- The promoter, the molecular agent that helps to initiate and modulate the desired change.
- The pathway regulator, which finds the appropriate point in the cell’s signaling pathway to make the intervention.
“The pieces that we used to build this control system existed,” Smolke said. “Combining them in a modular fashion into this molecular network diverter is what’s new.”
Inducing or inhibiting yeast mating
By chemically tuning this molecular network diverter in different ways, the researchers were able to: induce yeast to mate in the absence of pheromones; inhibit mating behavior even when pheromones were present; and engineer cell lines that contained this pathway-regulating mechanism in a neutral state until and unless researchers flipped this chemical switch.
“We’ve created this tool, and the tool itself is the control system and the methodology of implementing it,” Smolke said, adding that based on this proof of principle “we’re able to propose other generalized design strategies.”
Her assessment was echoed by James Collins, PhD, a professor of biomedical engineering at Boston University and a researcher with the Wyss Institute at Harvard University.
Collins, who was not associated with the experiment, said the paper demonstrated a new, systems-approach toward building tools for bioengineering.
“It shows that we don’t need to start from scratch. We can achieve advances by harnessing components that we already have,” said Collins, who described the work as “an application of engineering principles to cell biology” and “an important advance for synthetic biology.”
The work was funded by the National Institutes of Health, the National Science Foundation, the U.S. Defense Advanced Research Projects Agency and the Bill and Melinda Gates Foundation.