Building ‘smart’ cell-based therapies
April 22, 2014
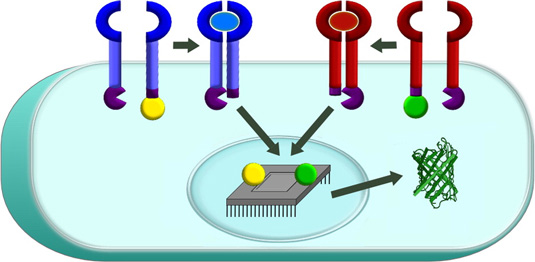
A technology for engineering human cell-based “devices” that monitor and modify human physiology. A protein biosensor sits on the surface of a cell, can be programmed to sense specific external factors, and upon detecting these factors sends a signal into the engineered cell’s nucleus to activate a gene expression program. (Credit: Northwestern’s McCormick School of Engineering and Applied Science)
A Northwestern synthetic biology team has created a new technology for modifying human cells to create programmable therapeutics that could travel the body and selectively target cancer and other sites of disease.
Engineering cell-based biological devices that monitor and modify human physiology is a promising frontier in clinical synthetic biology. However, no existing technology has enabled bioengineers to build devices that sense a patient’s physiological state and respond in a customized fashion.
Programmable biosensors
“The project addressed a key gap in the synthetic biology toolbox,” says Joshua Leonard, assistant professor of chemical and biological engineering in Northwestern’s McCormick School of Engineering and Applied Science. “There was no way to engineer cells in a manner that allowed them to sense key pieces of information about their environment, which could indicate whether the engineered cell is in healthy tissue or sitting next to a tumor.”
Leonard’s team worked for nearly four years to close this gap. The end result is a protein biosensor that sits on the surface of a cell and can be programmed to sense specific external factors. For example, the engineered cell could detect big, soluble protein molecules that indicate that it’s next to a tumor.
When the biosensor detects such a factor, it sends a signal into the engineered cell’s nucleus to activate a gene expression program, such as the production of tumor-killing proteins or chemicals. Since this toxic program would be activated only near tumor cells, such an approach could minimize side effects as well as improve therapeutic benefits.
Called a Modular Extracellular Sensor Architecture (MESA), the biosensor platform is completely self-contained so that several different biosensors can be present in a single cell without interfering with one another, allowing bioengineers to build increasingly sophisticated functional programs. The platform is also highly modular, enabling the biosensors to be customized to recognize factors of relevance to various patients’ needs.
Logic operations
“By linking the output of these biosensors to genetic programs, one can build in a certain logical command, such as ‘turn the output gene on when you sense this factor but not that factor,’” Leonard explains. “In that way, you could program a cell-based therapy to specify which cells it should kill.”
Leonard says doctors could potentially collect immune cells from a patient’s body, engineer the cells using MESA, and put them back into the patient. From there, the cells would do the work of detecting cancer or the disease they are designed to identify.
This is the first completely ground-up engineering of a receptor, and now that the core technology has been established, Leonard’s team is moving forward to program cells to recognize specific tumor-associated factors. They are also looking toward applications beyond advanced cell-based therapies.
Funded by the National Academies Keck Futures Initiative and the Defense Advanced Research Projects Agency, the research is published online in the journal ACS Synthetic Biology.
Abstract of ACS Synthetic Biology paper
Engineering mammalian cell-based devices that monitor and therapeutically modulate human physiology is a promising and emerging frontier in clinical synthetic biology. However, realizing this vision will require new technologies enabling engineered circuitry to sense and respond to physiologically relevant cues. No existing technology enables an engineered cell to sense exclusively extracellular ligands, including proteins and pathogens, without relying upon native cellular receptors or signal transduction pathways that may be subject to crosstalk with native cellular components. To address this need, we here report a technology we term a Modular Extracellular Sensor Architecture (MESA). This self-contained receptor and signal transduction platform is maximally orthogonal to native cellular processes and comprises independent, tunable protein modules that enable performance optimization and straightforward engineering of novel MESA that recognize novel ligands. We demonstrate ligand-inducible activation of MESA signaling, optimization of receptor performance using design-based approaches, and generation of MESA biosensors that produce outputs in the form of either transcriptional regulation or transcription-independent reconstitution of enzymatic activity. This systematic, quantitative platform characterization provides a framework for engineering MESA to recognize novel ligands and for integrating these sensors into diverse mammalian synthetic biology applications.