Craig Venter’s team designs, builds first minimal synthetic bacterial cell
March 28, 2016
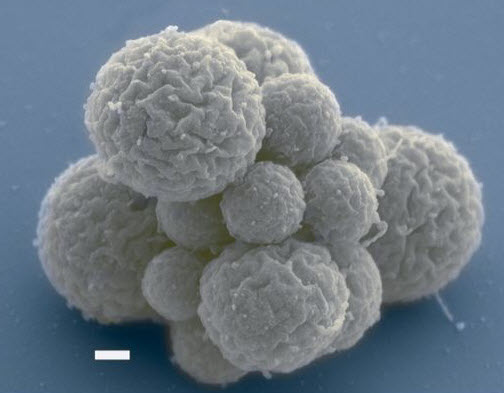
A cluster of cells of a new synthetic species known as JCVI-syn3.0, showing spherical structures of varying sizes (scale bar, 200 nm). (credit: Clyde A. Hutchison III et al./Science)
Just 473 genes were needed to create life in a new synthesized species of bacteria created by synthetic biologists from the J. Craig Venter Institute (JCVI) and Synthetic Genomics, Inc.
Knowing the minimum number of genes to create life would answer a fundamental question in biology.
This “minimal synthetic cell,” JCVI-syn3.0, was reported in an open-access paper published last week in the journal Science. By comparison, the first synthetic cell developed by the scientists, M. mycoides JCVI-syn1.0, has 1.08 million base pairs and 901 genes.*
The new cell contains 531,560 base pairs (the “alphabet” or sequence that makes up the DNA code) and 473 genes — the smallest number of genes of any organism that can be grown in a laboratory, according to the team.
JCVI | CVI-syn3.0 — Minimal Cell
“All of the…studies over the past 20 years have underestimated the number of essential genes by focusing only on the known world. This is an important observation that we are carrying forward into the study of the human genome,” said senior author and group leader J. Craig Venter, PhD.
For 50 years, researchers have studied essential and non-essential genes in bacteria to help biologists understand the core functions needed for life. In the newer field of synthetic biology, this same information will be able to help scientists design DNA sequences for new synthetic organisms — allowing them to build frameworks for industrial applications of synthetic organisms.
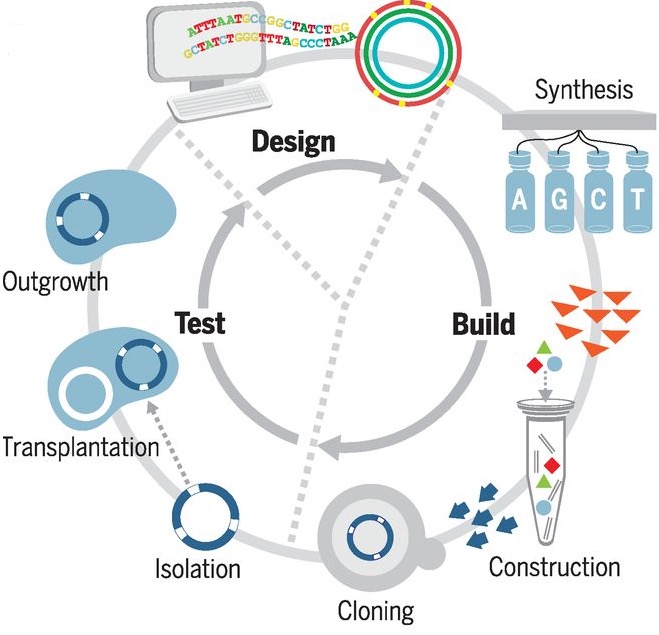
The cycle for genome design, building by means of synthesis and cloning in yeast, and testing for viability by means of genome transplantation. After each cycle, gene essentiality is reevaluated by global transposon mutagenesis. (credit: Clyde A. Hutchison III et al./Science)
Mystery genes
During construction** of JCVI-syn3.0, the team discovered that 149 of the genes actually had unknown functions, but were essential for robust growth. Those functions remain an area of continued work for the researchers. Other genes in the minimal synthetic cell were related to reading and expressing the DNA code, structure, and function of the outer cell membrane, and cell metabolism, or to preserving DNA integrity.
However, the team expects to decode the 149 genes in the future. “Our goal is to have a cell for which the precise biological function of every gene is known,” said Clyde Hutchison, PhD, first author and distinguished professor at JCVI.
Another major outcome of the minimal cell program will be new tools and semi-automated processes for synthesizing the DNA sequence needed for whole organisms, according to Daniel Gibson, PhD, an associate professor at JCVI.
“This paper signifies a major step toward our ability to design and build synthetic organisms from the bottom up with predictable outcomes,” he said. “The tools and knowledge gained from this work will be essential to producing next-generation production platforms for a wide range of disciplines.”
This work was funded by SGI, the JCVI endowment, and the Defense Advanced Research Projects Agency’s Living Foundries program.
* The research at JCVI leading to this report began in 1995 with DNA sequencing of the first free-living organism, Haemophilus influenza, followed by the DNA sequencing of Mycoplasma genitalium. A comparison of these two genomes revealed a common set of 256 genes that the team thought could be a minimal set of genes needed for viability.
In 1999, Hutchison led a team who published a paper describing techniques to identify the non-essential genes in M. genitalium.
The creation of the first synthetic cell (JCVI-syn1.0) in 2010 established a workflow for building and testing designs for the DNA of a whole organism. This included design of a minimal cell from the bottom up, starting with the DNA sequence.
** To create JCVI-syn3.0, the team used an approach of whole genome design (design of the DNA needed for a whole organism) and chemical synthesis followed by genome transplantation to test if the cell was viable. Their first attempt to minimize the genome began with a simple approach using information in the biochemical literature and some limited mutations of DNA, but this did not result in a viable genome. After improving methods, the team discovered a set of “quasi-essential” genes that are necessary for robust growth and that explained the failure of their first attempt.
The team built the genome in eight segments at a time so that each could be tested separately before combining them to generate a minimal genome. The team also explored the order of the genes and how that affects cell growth and viability. They found gene content was more critical than gene order. They went through three cycles of designing, building, and testing ensuring that the “quasi-essential” genes remained, which in the end resulted in a viable, self-replicating minimal synthetic cell that contained just 473 genes.
Abstract of Design and synthesis of a minimal bacterial genome
We used whole-genome design and complete chemical synthesis to minimize the 1079–kilobase pair synthetic genome of Mycoplasma mycoides JCVI-syn1.0. An initial design, based on collective knowledge of molecular biology combined with limited transposon mutagenesis data, failed to produce a viable cell. Improved transposon mutagenesis methods revealed a class of quasi-essential genes that are needed for robust growth, explaining the failure of our initial design. Three cycles of design, synthesis, and testing, with retention of quasi-essential genes, produced JCVI-syn3.0 (531 kilobase pairs, 473 genes), which has a genome smaller than that of any autonomously replicating cell found in nature. JCVI-syn3.0 retains almost all genes involved in the synthesis and processing of macromolecules. Unexpectedly, it also contains 149 genes with unknown biological functions. JCVI-syn3.0 is a versatile platform for investigating the core functions of life and for exploring whole-genome design.