Deep Genomics launches, uniting deep learning and genome biology
July 22, 2015
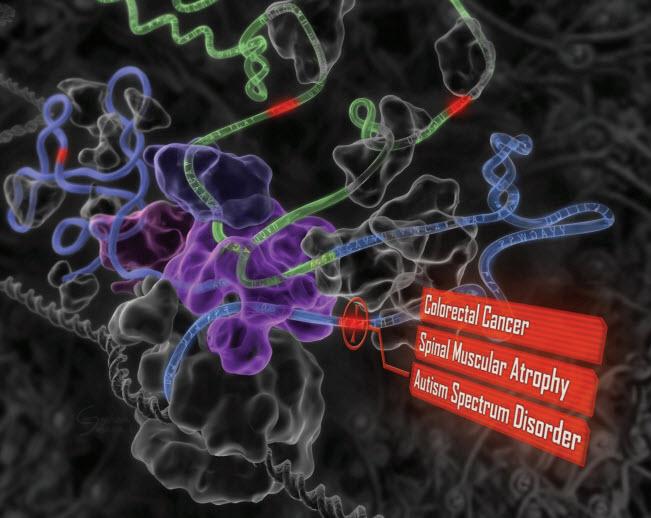
“Deep learning” reveals the genetic origins of disease. A computational system mimics the biology of RNA splicing by correlating DNA elements with splicing levels in healthy human tissues. The system can scan DNA and identify damaging genetic variants, including those deep within introns.This procedure has led to insights into the genetics of autism, cancers, and spinal muscular atrophy. (credit: Hui Y. Xiong et al./Science)
Deep Genomics, a University of Toronto spinoff, launched today (July 22), combining deep learning and artificial intelligence with the study of the human genome. The company is building on more than a decade of research and expertise in both fields.
Using deep learning allows Deep Genomics to predict the consequences of genomic alteration on various cell mechanisms to make life-changing decisions, potentially via personalized medicine treatment, the researchers say.
“Our vision is to change the course of genomic medicine and help save lives by determining smarter treatment options,” says Brendan Frey, the company’s president and CEO, a Fellow of the Canadian Institute for Advanced Research, and a Professor at the University of Toronto.
SPIDEX, a Google for DNA mutation effects
Professor Brendan Frey (center-right) and colleagues at the University of Toronto Faculty of Applied Science & Engineering (credit: Roberta Baker/ U of T Engineering)
The scientific community has spent decades searching for mutations within specific genes that can be connected to disease, such as the BRCA-1 and BRCA-2 genes for breast cancer. But there is a vast amount of mutations and combinations of mutations that have neither been observed nor studied, posing a challenge for diagnostics and therapeutics today.
“We envision a future where computers are trusted to predict the outcome of laboratory experiments and treatments, long before anyone picks up a test tube. Our first step will be to open up a genome-wide database of over 300 million potentially disease-causing variants, most of which are in regions of the genome that can’t be examined using other methods.”
Deep Genomics’ first product, called SPIDEX, provides information about how these DNA mutations may alter splicing in the cell, a process that is crucial for normal development. It also connects the dots between a variant or mutation of unknown significance and a variant that has been linked to a disease to determine its level of danger.
Because errant splicing is behind many diseases and disorders, including cancers and autism spectrum disorder, SPIDEX has immediate and practical importance for genetic testing and pharmaceutical development. The science validating the SPIDEX tool was described in the January 9, 2015 issue of the journal Science.
Labs will send the mutations they’ve collected to Deep Genomics, and the company will use their proprietary deep learning system, which includes SPIDEX, to “read” the genome and assess how likely the mutation is to cause a problem. SPIDEX can also connect the dots between a variant of unknown significance and a variant that has been linked to disease.
The company plans to further grow its team of machine learning, genome biology, and computational biology experts, and continue to invent new deep learning technologies and work with diagnosticians and biologists to understand the many complex ways that cells interpret DNA.
The company’s scientific advisory board includes Yann LeCun, Director, Facebook AI Research; Stephen Scherer, Director, The Center for Applied Genomics; and Jordan Lerner-Ellis, Director, Molecular Diagnostics at Mount Sinai Hospital.
More information: www.deepgenomics.com.
Abstract of The human splicing code reveals new insights into the genetic determinants of disease
To facilitate precision medicine and whole-genome annotation, we developed a machine-learning technique that scores how strongly genetic variants affect RNA splicing, whose alteration contributes to many diseases. Analysis of more than 650,000 intronic and exonic variants revealed widespread patterns of mutation-driven aberrant splicing. Intronic disease mutations that are more than 30 nucleotides from any splice site alter splicing nine times as often as common variants, and missense exonic disease mutations that have the least impact on protein function are five times as likely as others to alter splicing. We detected tens of thousands of disease-causing mutations, including those involved in cancers and spinal muscular atrophy. Examination of intronic and exonic variants found using whole-genome sequencing of individuals with autism revealed misspliced genes with neurodevelopmental phenotypes. Our approach provides evidence for causal variants and should enable new discoveries in precision medicine.