Designing interlocking building blocks to create complex tissues
March 13, 2013
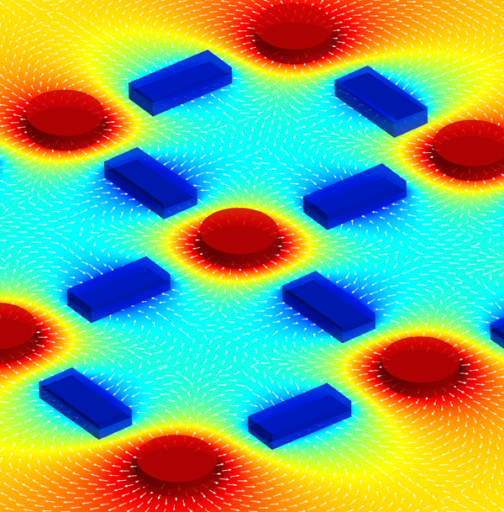
This image illustrates mathematical modeling of the migration of mesenchymal stem cells (encapsulated in cylinders) in response to signals released by endothelial cells (encapsulated in rectangles). The color intensity corresponds to concentration, and the arrows represent directions of cell motion. (Credit: George Eng/Columbia University School of Engineering and Applied Science)
Columbia University researchers have developed a new “plug-and-play” method to assemble complex cell microenvironments in a scalable, highly precise way to fabricate tissues with any spatial organization or interest — such as those found in the heart or skeleton or vasculature.
The lock-and-key technique can be used to build cellular assemblies using a variety of shapes that lock into templates like LEGO building blocks, according to Gordana Vunjak-Novakovic, who led the study and is the Mikati Foundation Professor of Biomedical Engineering at Columbia Engineering and professor of medical sciences.
“What is really important about this technique is that these shapes are tiny — just a fraction of a millimeter, the thickness of a human hair — and that their precise arrangements are made using cell-friendly hydrogels.”
Tissue cells in the human body form specific architectures that are critical for the function of each tissue. Cardiac cells, for example, are aligned to create maximum force acting in one direction. Cells without specific spatial organization may never become fully functional if they do not recapitulate their intrinsic organization found in the body.
The Columbia Engineering technique enables researchers to construct unique and controlled cell patterns that allow precise studies of cell function. “we can now ask some of the more complex questions about how the cells respond to the entire context of their environment,” says Vunjak-Novakovic. This will help us explore cellular behavior during the progression of disease and test the effects of drugs, stem cells, and various other therapeutic measures.”
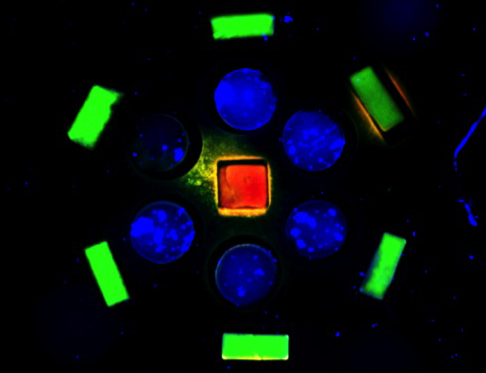
Shapes in their congruent wells after sorting (credit: George Eng et al./Columbia University School of Engineering and Applied Science)
Computer-chip fab techniques
The researchers used computer-chip-fabrication technologies to make micron-sized subunits containing living cells and bioactive molecules, in specific building-block geometries, such as cylinders or cubes.
Each shape is prepared to have its own unique biological properties and is then placed into its geometrically matching well on the hydrogel template. The assembling technique is simple: a mixture of various types of shapes is pipetted onto a template with a specific arrangement of the matching wells and placed on a laboratory shaker for a few minutes.
Each shape can dock only into its matching well — the rectangular blocks dock only into the exact same rectangular wells, cylindrical blocks into cylindrical wells, and so on, in a lock-and-key manner. After up to ten short cycles of shaking, the template becomes filled with shapes to form a precisely defined pattern. This docking technique thus allows rapid assembly of a large number of subunits to create new tissues.
“The beauty of this method is that complex configurations of living cellular material — many different types of cells, molecules, and extracellular materials — emerge in the lab in precise three-dimensional geometries in a way that can be used by anyone, as no special equipment is involved,” says Vunjak-Novakovic.
Next steps in the application of this new technique include fabrication of different types of functional tissues, such as well-organized cardiac muscle, a tissue whose function critically depends on its architecture and cell alignment, incorporating blood vessel networks along with organized cardiac cells. The method will also be extended to the design of pathological microenvironments of interest, such as tumor models.
This research is supported by the National Institutes of Health, the Helmsley Foundation, and Columbia’s MD/PhD program.