Engineered bacteria form multicellular circuit to control protein expression
August 31, 2015
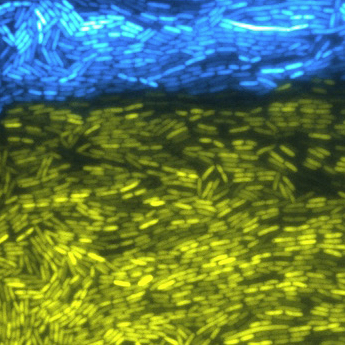
Two strains of synthetically engineered bacteria cooperate to create multicellular phenomena. Their fluorescence indicates the engineered capabilities have been activated. (credit: Bennett Lab/Rice University)
Rice University scientists and associates have created a biological equivalent to a computer circuit using multiple types of bacteria that change protein expression. The goal is to modify biological systems by controlling how bacteria influence each other. This could lead to bacteria that, for instance, beneficially alter the gut microbiome (collection of microorganisms) in humans.
The research is published in the journal Science.
Humans’ stomachs have a lot of different kinds of bacteria contained in the microbiome. “They naturally form a large consortium,” said Rice synthetic biologist Matthew Bennett. The idea is to engineer bacteria to be part of a consortium. “Working together allows them to effect more change than if they worked in isolation.”
In the proof-of-concept study, Bennett and his team created two strains of genetically engineered bacteria that regulate the production of proteins essential to intercellular signaling pathways, which allow cells to coordinate their efforts, generally in beneficial ways.
The synthetic microbial consortium oscillator yo-yo
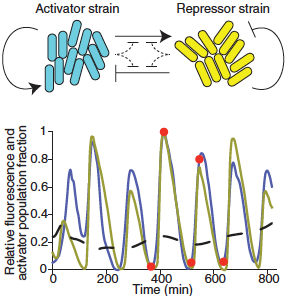
The activator strain up-regulates genes in both strains; the repressor strain down-regulates genes in both strains, generating an oscillation of gene transcription in the bacterial population (credit: Ye Chen et al.)
“The main push in synthetic biology has been to engineer single cells,” Bennett said. “But now we’re moving toward multicellular systems. We want cells to coordinate their behaviors in order to elicit a populational response, just the way our bodies do.”
Bennett and his colleagues achieved their goal by engineering common Escherichia coli bacteria. By creating and mixing two genetically distinct populations, they prompted the bacteria to form a consortium.
The bacteria worked together by doing opposite tasks: One was an activator that up-regulated the expression of targeted genes; the other was a repressor that down-regulated specific genes. Together, they created oscillations of gene transcription in the bacterial population.
The two novel strains of bacteria sent out intercellular signaling molecules and created linked positive and negative feedback loops that affected gene production in the entire population. Both strains were engineered to make fluorescent reporter genes so their activities could be monitored. The bacteria were confined to microfluidic devices in the lab, where they could be monitored easily during each hours-long experiment.
When the bacteria were cultured in isolation, the protein oscillations did not appear, the researchers wrote.
Programmed yogurt, anyone?
Bennett said his lab’s work will help researchers understand how cells communicate, an important factor in fighting disease. “We have many different types of cells in our bodies, from skin cells to liver cells to pancreatic cells, and they all coordinate their behaviors to make us work properly,” he said. “To do this, they often send out small signaling molecules that are produced in one cell type and effect change in another cell type.
“We take that principle and engineer it into these very simple organisms to see if we can understand and build multicellular systems from the ground up.”
Ultimately, people might ingest the equivalent of biological computers that can be programmed through one’s diet, Bennett said. “One idea is to create a yogurt using engineered bacteria,” he said. “The patient eats it and the physician controls the bacteria through the patient’s diet. Certain combinations of molecules in your food can turn systems within the synthetic bacteria on and off, and then these systems can communicate with each other to effect change within your gut.”
KAIST and University of Houston scientists were also involved in the research. The National Institutes of Health, the Robert A. Welch Foundation, the Hamill Foundation, the National Science Foundation, and the China Scholarship Council supported the research.
Abstract of Emergent genetic oscillations in a synthetic microbial consortium
A challenge of synthetic biology is the creation of cooperative microbial systems that exhibit population-level behaviors. Such systems use cellular signaling mechanisms to regulate gene expression across multiple cell types. We describe the construction of a synthetic microbial consortium consisting of two distinct cell types—an “activator” strain and a “repressor” strain. These strains produced two orthogonal cell-signaling molecules that regulate gene expression within a synthetic circuit spanning both strains. The two strains generated emergent, population-level oscillations only when cultured together. Certain network topologies of the two-strain circuit were better at maintaining robust oscillations than others. The ability to program population-level dynamics through the genetic engineering of multiple cooperative strains points the way toward engineering complex synthetic tissues and organs with multiple cell types.