How to create models of biomolecules using x-rays
May 29, 2013

Fluctuation x-ray scattering is the basis of a new technique for rapidly modeling the shapes of large biological models, here demonstrated (gray envelopes) using existing diffraction data superposed on known high-resolution structures. Top left, lysine-arginine-ornithine (LAO) binding protein; top right, lysozome; bottom left, peroxiredoxin; and, bottom right, Satellite Tobacco Mosaic Virus (STMV). (Credit: Lawrence Berkeley National Laboratory)
Berkeley Lab researchers and their colleagues have created a new way to model biological molecules using x-rays.
Existing methods for solving structure largely depend on crystallized molecules, and the shapes of more than 80,000 proteins in a static state have been solved this way.
Most of the two million proteins in the human body can’t be crystallized, however, so even their low-resolution structures are still unknown.
Now, new techniques developed by Peter Zwart and colleagues at the U.S. Department of Energy’s Lawrence Berkeley National Laboratory (Berkeley Lab) and collaborators (DOE’s Pacific Northwest National Laboratory (PNNL), Arizona State University, and the University of Wisconsin-Milwaukee) promise a better look at large biological molecules in their native, more-fluid state.
Diffraction before destruction
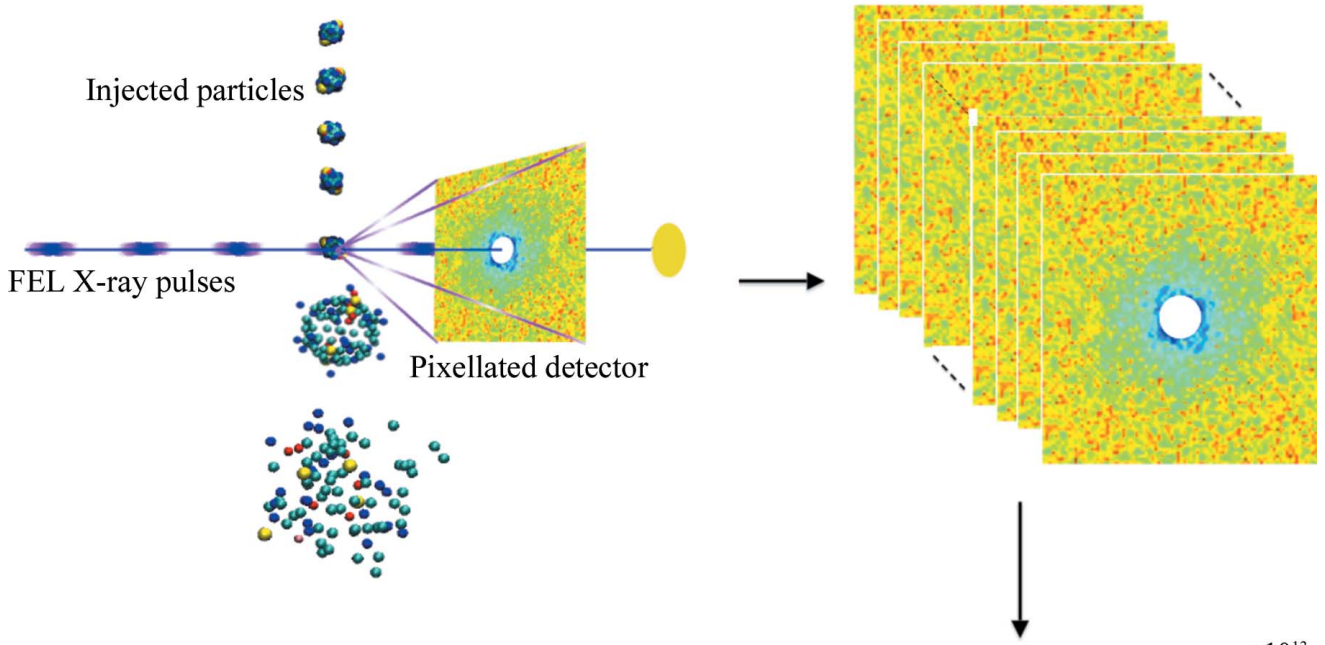
“Diffract before destroy” technique: numerous copies of a molecule in solution are fired across the x-ray beam of a free-electron laser. Diffraction patterns are collected just before the femtosecond x-ray pulses explode the particles…. (Credit: Lawrence Berkeley National Laboratory)
It uses data created by free-electron lasers (FELs) such as the Linac Coherent Light Source (LCLS) at SLAC National Accelerator Laboratory, or the proposed Next Generation Light Source (NGLS), light sources whose powerful x‑ray pulses are measured in quadrillionths of a second.
These pulses are faster than a molecule can rotate, so the experimental data reflects the state of the molecule frozen in time.
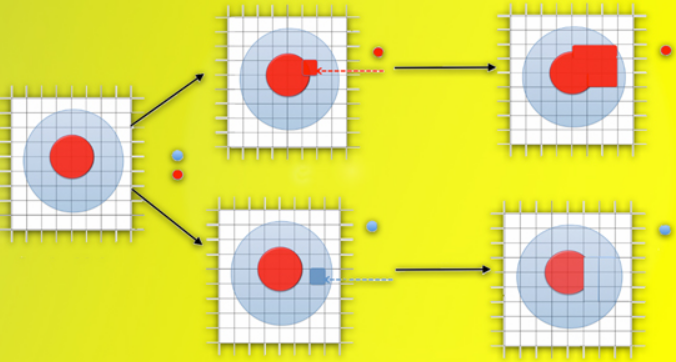
… A 3D model is then constructed by repeatedly enlarging and removing parts of an arbitrary shape until it closely matches the average values derived from the diffraction patterns (credit: Lawrence Berkeley National Laboratory)
“It’s a technique called ‘diffract before destroy,’ because the data is collected before the particle literally blows apart,” says Zwart, a member of the Lab’s Physical Biosciences Division, and the science lead for the Berkeley Center for Structural Biology at the Advanced Light Source.
“FELs have shown they can derive structures from single particles, each hit with a single pulse, but there are major challenges to this approach.”
Instead of single particles, Zwart and his colleagues include many particles in each shot.
When analyzed by computer programs, the data from the different diffraction patterns can be combined to provide detailed insights into the structures the molecules adopt in solution.
Fluctuation x-ray scattering
The technique is an example of fluctuation x-ray scattering (fXS), and Zwart and his colleagues have shown that data obtained this way with free-electron lasers can yield low-resolution shapes of biomolecules in close to their natural state, with much greater confidence than is currently possible with less powerful synchrotron light sources.
“Our algorithm starts with a trial model and modifies it by randomly adding or subtracting volume until the shape of the model achieves the optimum fit with the data,” Zwart says. This trial-and-error optimization technique, tested on known configurations at the LCLS, can resolve the shapes of individual macromolecules with fXS data alone.
Zwart and his former postdoc Gang Chen, working with Dongsheng Li of PNNL, have also shown that data from mixtures of different kinds of molecules can be untangled to provide clues on the structure of the individual components, forming a basis for understanding the dynamic behavior of large biological molecules working together in solution.
By understanding their structural changes, Zwart and his colleagues are developing fluctuation x-ray scattering as an indispensable tool for determining how mixtures of different proteins behave independently or in concert.
This work was supported by DOE’s Office of Science through Laboratory Directed Research and Development programs at Berkeley Lab and PNNL; the international Human Frontier Science Program; the National Science Foundation; the University of Wisconsin Research Growth Initiative; and the Chinese Academy of Sciences.