How to design proteins from scratch
November 8, 2012
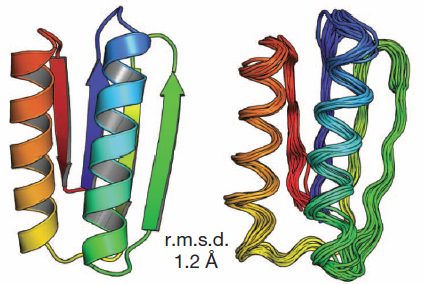
Comparison of computational models with experimentally
determined structure: design model (left) and NMR structure (right). Credit: Nobuyasu Koga et al./Nature)
Given the exponential number of contortions possible for any chain of amino acids, dictating a sequence that will fold into a predictable protein structure has been a daunting task.
Now a team from David Baker’s laboratory at the University of Washington reports that they can do just that, Nature News reports.
By following a set of rules, they designed five proteins from scratch that fold reliably into predicted conformations. In a blind test, the team showed that the synthesized proteins closely match the predicted structures.
“What you have now is a flexible set of building blocks for nanoscale assembly,” says Jeremy England, a molecular biophysicist at the Massachusetts Institute of Technology in Cambridge, who was not involved in the work.
The work was spearheaded by husband-and-wife team Nobuyasu Koga and Rie Tatsumi-Koga, protein engineers in Baker’s group. After observing the backbone structures of thousands of proteins, they developed some intuitive rules they wanted to test.
Protein strands typically form helices and other classic secondary structures that in turn fold into the final protein shape. The team realized that these structures could be made to twist in one direction or another depending on the length of the loops that connected them. By choosing the right sequence lengths between these building blocks, the team could predict which way they would fold.