How to sort and extract biomolecules in fluids
March 24, 2015
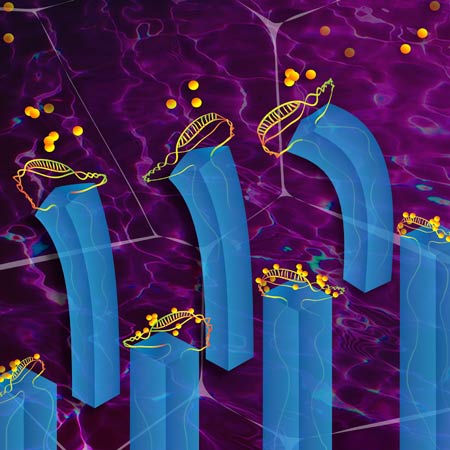
Detecting and extracting biomolecules from fluid mixtures (credit: Peter Mallen, Harvard Medical School)
Harvard scientists have demonstrated a new way to detect and extract biomolecules from fluid mixtures, using an ingenious microfluidic design combining chemical and mechanical properties.
The approach requires fewer steps, uses less energy, and achieves better performance than several techniques currently in use. It could lead to better technologies for medical diagnostics and chemical purification.
For example, it could provide a means of removing contaminants from water, and even be tailored to enable energy-efficient desalination of seawater, the researchers say. It could also be used to capture valuable minerals from fluid mixtures.
The biomolecule sorting technique was developed in the laboratory of Joanna Aizenberg, Amy Smith Berylson Professor of Materials Science at Harvard School of Engineering and Applied Sciences (SEAS) and Professor in the Department of Chemistry and Chemical Biology.
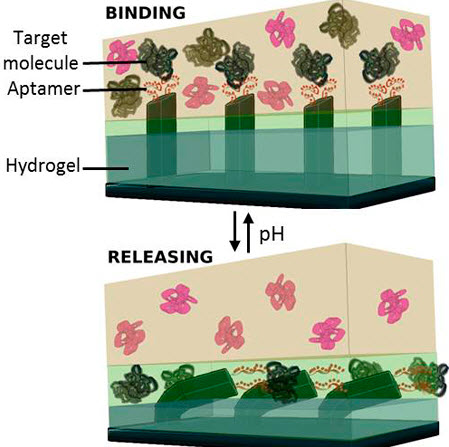
Capture and release of specific target biomolecules from an ingoing solution mixture in a microfluidic system occurs by the concerted, dynamic and reversible action of hydrogel volume change and aptamer bind-and-release through changes in solution pH. (credit: Ankita Shastri and Ximin He)
How it works
The new microfluidic device, described in a paper appearing Monday March 23 in the journal Nature Chemistry, is composed of microscopic “fins” embedded in a hydrogel that is able to respond to different stimuli, such as temperature, pH, and light.
Special DNA strands called aptamers, that under the right conditions bind to a specific target molecule, are attached to the fins, which move the cargo between two chemically distinct environments. Modulating the pH levels of the solutions in those environments triggers the aptamers to “catch” or “release” the target biomolecule.
After using computer simulations to test their novel approach, in collaboration with Prof. Anna C. Balazs from the University of Pittsburgh, Aizenberg’s team conducted proof-of-concept experiments in which they successfully separated thrombin, an enzyme in blood plasma that causes the clotting of blood, from several mixtures of proteins.
Their research suggests that the technique could be applicable to other biomolecules, or used to determine chemical purity and other characteristics in inorganic and synthetic chemistry.
“Our adaptive hybrid sorting system presents an efficient chemo-mechanical transductor, capable of highly selective separation of a target species from a complex mixture — all without destructive chemical modifications and high-energy inputs,” Aizenberg said. “This new approach holds promise for next-generation, energy-efficient separation and purification technologies and medical diagnostics.”
Tunable, repeatable, reusable
The system is dynamic; its integrated components are highly tunable. For example, the chemistry of the hydrogel can be modified to respond to changes in temperature, light, electric and magnetic fields, and ionic concentration. Aptamers, meanwhile, can target a range of proteins and molecules in response to variations in pH levels, temperature, and salt.
“The system allows repeated processing of a single input solution, which enables multiple recycling and a high rate of capture of the target molecules,” said lead author Ximin He, Assistant Professor of Materials Science and Engineering at Arizona State University and formerly a postdoctoral research fellow in Aizenberg’s group at Harvard.
Conventional biomolecule sorting systems rely on external electric fields, infrared radiation, and magnetic fields, and often require chemical modifications of the biomolecules of interest. That means setups can be used only once or require a series of sequential steps.
Aizenberg is also co-director of the Kavli Institute for Bionano Science and Technology and a core faculty member at Harvard’s Wyss Institute for Biologically Inspired Engineering, leading the Adaptive Materials Technologies platform there.
Other contributors to the work include researchers from the University of Pittsburgh, Arizona State University, and the Wyss Institute. The research was supported by the U.S. Department of Energy.
Abstract of An aptamer-functionalized chemomechanically modulated biomolecule catch-and-release system
The efficient extraction of (bio)molecules from fluid mixtures is vital for applications ranging from target characterization in (bio)chemistry to environmental analysis and biomedical diagnostics. Inspired by biological processes that seamlessly synchronize the capture, transport and release of biomolecules, we designed a robust chemomechanical sorting system capable of the concerted catch and release of target biomolecules from a solution mixture. The hybrid system is composed of target-specific, reversible binding sites attached to microscopic fins embedded in a responsive hydrogel that moves the cargo between two chemically distinct environments. To demonstrate the utility of the system, we focus on the effective separation of thrombin by synchronizing the pH-dependent binding strength of a thrombin-specific aptamer with volume changes of the pH-responsive hydrogel in a biphasic microfluidic regime, and show a non-destructive separation that has a quantitative sorting efficiency, as well as the system’s stability and amenability to multiple solution recycling.