Molecules from scratch without the fiendish physics
February 12, 2012 | Source: New Scientist
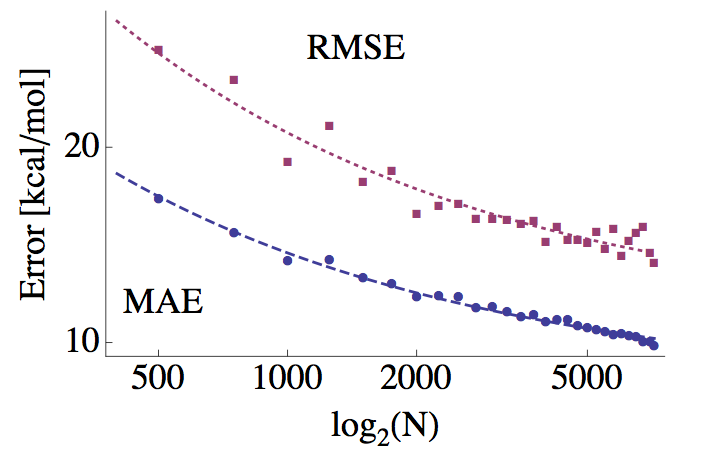
Cross-validated machine learning errors as a function of number of molecules in training set, N (credit: Matthias Rupp et al./PRL)
A suite of artificial intelligence algorithms developed at Argonne National Laboratory can now predict a property of molecules from their theoretical structure in a millisecond that would take earlier methods an hour.
Similar advances should allow chemists to design new molecules on computers instead of by lengthy trial-and-error.
They bypassed the complex Schrödinger equation entirely by building a database of 7165 molecules with known atomization energies and structures. They used 1000 machine learning techniques able to predict atomization energies from patterns in large data sets with complicated underlying rules.
Ref.: Matthias Rupp et al., Fast and Accurate Modeling of Molecular Atomization Energies with Machine Learning, Physical Review Letters, 2012 [DOI: 10.1103/PhysRevLett.108.058301]
Ref.: Matthias Rupp et al., Fast and Accurate Modeling of Molecular Atomization Energies with Machine Learning, arxiv.org/abs/1109.2618