New protein structure could help diagnose and treat Alzheimer’s, many related diseases
August 7, 2014
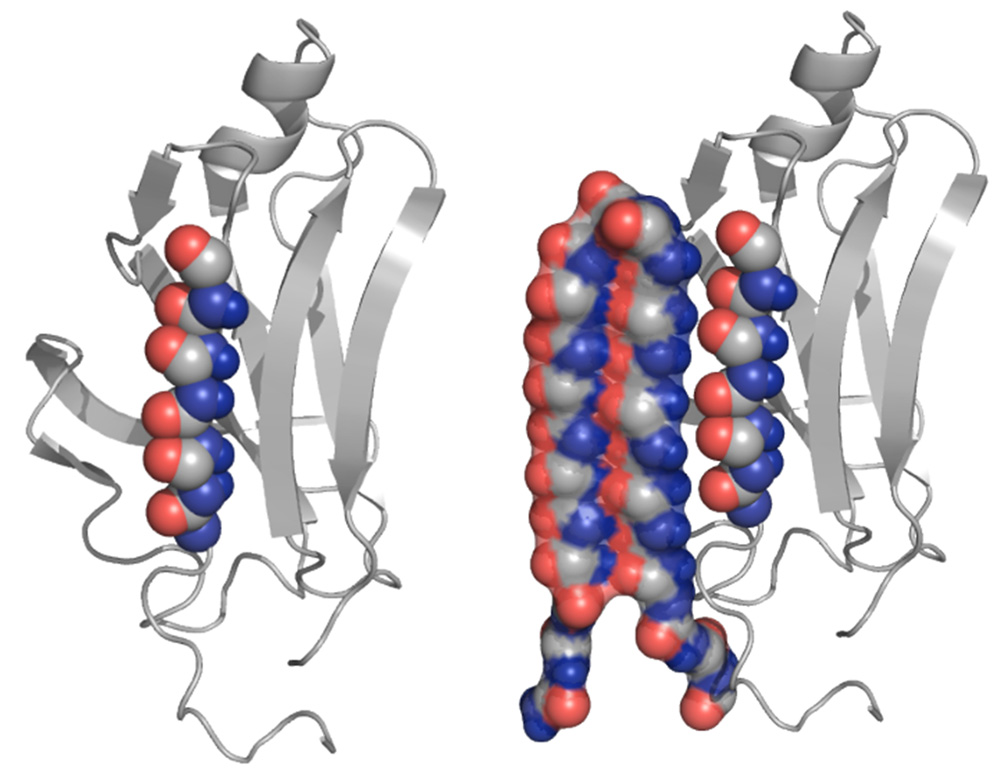
An abnormal protein, left, is intercepted by the UW’s compound, which can bind to the toxic protein and neutralize it, as shown at right (credit: University of Washington)
University of Washington (UW) bioengineers have designed a peptide structure that they say could stop the harmful changes of the body’s proteins linked to widespread diseases such as Alzheimer’s, Parkinson’s, heart disease, Type 2 diabetes, and Lou Gehrig’s disease.
The new synthetic molecule blocks these proteins as they shift from their normal state into an abnormally folded form, by targeting a toxic intermediate phase.
The discovery of a protein blocker could lead to ways to diagnose and even treat a large swath of diseases that are hard to pin down and rarely have a cure.
The findings were published in the journal eLife (open access).
“If you can truly catch and neutralize the toxic version of these proteins, then you hopefully never get any further damage in the body,” said senior author Valerie Daggett, a UW professor of bioengineering. “What’s critical with this and what has never been done before is that a single peptide sequence will work against the toxic versions of a number of different amyloid proteins and peptides, regardless of their amino acid sequence or the normal 3D structures.”
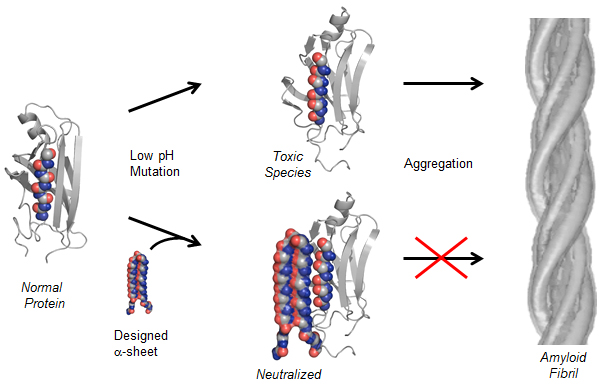
In this diagram, a normal protein begins to convert into a toxic, intermediate state (above center). The UW’s compound can bind with the toxic species and neutralize it (below center), preventing amyloid fibrils (right) from forming. (Credit: University of Washington)
More than 40 illnesses known as amyloid diseases — Alzheimer’s, Parkinson’s and rheumatoid arthritis are a few — are linked to the buildup of proteins after they have transformed from their normally folded, biologically active forms to abnormally folded, grouped deposits called fibrils or plaques.
This happens naturally as we age, to a certain extent — our bodies don’t break down proteins as quickly as they should, causing higher concentrations in some parts of the body.
Each amyloid disease has a unique, abnormally folded protein or peptide structure, but often such diseases are misdiagnosed because symptoms can be similar; and pinpointing which protein is present usually isn’t done until after death, in an autopsy.
As a result, many dementias are broadly diagnosed as Alzheimer’s disease without definitive proof, and other diseases can go undiagnosed and untreated.
The molecular structure of an amyloid protein can be only slightly different from a normal protein and can transform to a toxic state fairly easily, which is why amyloid diseases are so prevalent.
The researchers built a protein structure, called “alpha sheet,” that complements the toxic structure of amyloid proteins that they discovered in computer simulations. The alpha sheet effectively attacks the toxic middle state (see illustration above) the protein goes through as it transitions from normal to abnormal.
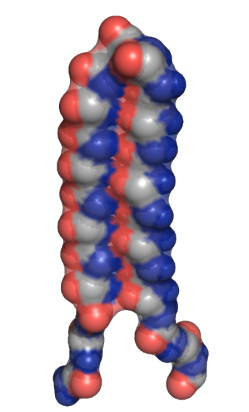
The UW’s designed “alpha sheet” protein structure (credit: University of Washington)
The structures could be tailored even further to bind specifically with the proteins in certain diseases, which could be useful for specific therapies.
The researchers hope their designed compounds could be used both as diagnostics for amyloid diseases and as drugs to treat the diseases or at least slow progression.
“For example, patients could have a broad first-pass test done to see if they have an amyloid disease and then drill down further to determine which proteins are present to identify the specific disease,” Daggett said.
Researchers at the National Institutes of Health’s Rocky Mountain Laboratories were also involved in the study.
The research was funded by the National Institutes of Health (General Medicine Sciences), the National Science Foundation, the Wallace H. Coulter Foundation, and Coins for Alzheimer’s Research Trust.
Abstract of eLife paper
Previous studies suggest that the toxic soluble-oligomeric form of different amyloid proteins share a common backbone conformation, but the amorphous nature of this oligomer prevents its structural characterization by experiment. Based on molecular dynamics simulations we proposed that toxic intermediates of different amyloid proteins adopt a common, nonstandard secondary structure, called α-sheet. Here we report the experimental characterization of peptides designed to be complementary to the α-sheet conformation observed in the simulations. We demonstrate inhibition of aggregation in two different amyloid systems, β-amyloid peptide (Aβ) and transthyretin, by these designed α-sheet peptides. When immobilized the α-sheet designs preferentially bind species from solutions enriched in the toxic conformer compared with non-aggregated, nontoxic species or mature fibrils. The designs display characteristic spectroscopic signatures distinguishing them from conventional secondary structures, supporting α-sheet as a structure involved in the toxic oligomer stage of amyloid formation and paving the way for novel therapeutics and diagnostics.