Periodic table of protein complexes helps predict novel protein structures
December 10, 2015
An interactive Periodic Table of Protein Complexes is available at http://sea31.user.srcf.net/periodictable/ (credit: EMBL-EBI/Spencer Phillips)
The Periodic Table of Protein Complexes, developed by researchers in the UK and to be published Dec. 11 in the journal Science, offers a new way of looking at the enormous variety of structures that proteins can build in nature. More importantly, it suggests which ones might be discovered next and how entirely novel structures could be engineered.
Created by an interdisciplinary team led by researchers at the Wellcome Genome Campus and the University of Cambridge, the Table provides a valuable tool for research into evolution and protein engineering.
Handling complexity
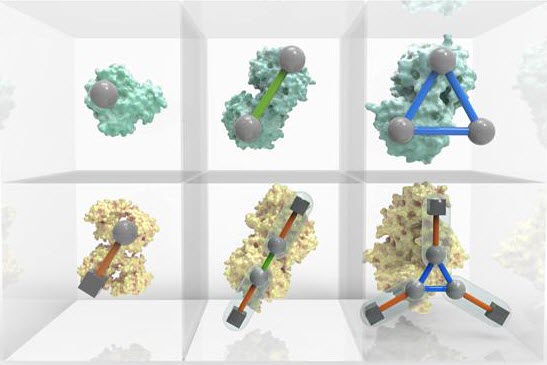
Almost every biological process depends on proteins interacting and assembling into complexes in a specific way, and many diseases are associated with problems in complex assembly. “Evolution has given rise to a huge variety of protein complexes, and it can seem a bit chaotic,” explains Joe Marsh of the MRC Human Genetics Unit at the University of Edinburgh. “But if you break down the steps proteins take to become complexes, there are some basic rules that can explain almost all of the assemblies people have observed so far.”
Fundamentally, protein complex assembly can be seen as endless variations on dimerization (one doubles, and becomes two), cyclisation (one forms a ring of three or more), and subunit addition (two different proteins bind to each other). Because these happen in a fairly predictable way, it’s not as hard as you might think to predict how a novel protein would form.
“By analyzing the tens of thousands of protein complexes for which three-dimensional structures have already been experimentally determined, we could see repeating patterns in the assembly transitions that occur — and with new data from mass spectrometry we could start to see the bigger picture,” says Marsh.
Abstract of Principles of assembly reveal a periodic table of protein complexes
INTRODUCTION: The assembly of proteins into complexes is crucial for most biological processes. The three-dimensional structures of many thousands of homomeric and heteromeric protein complexes have now been determined, and this has had a broad impact on our understanding of biological function and evolution. Despite this, the organizing principles that underlie the great diversity of protein quaternary structures observed in nature remain poorly understood, particularly in comparison with protein folds, which have been extensively classified in terms of their architecture and evolutionary relationships.
RATIONALE: In this work, we sought a comprehensive understanding of the general principles underlying quaternary structure organization. Our approach was to consider protein complexes in terms of their assembly. Many protein complexes assemble spontaneously via ordered pathways in vitro, and these pathways have a strong tendency to be evolutionarily conserved. Furthermore, there are strong similarities between protein complex assembly and evolutionary pathways, with assembly pathways often being reflective of evolutionary histories, and vice versa. This suggests that it may be useful to consider the types of protein complexes that have evolved from the perspective of what assembly pathways are possible.
RESULTS: We first examined the fundamental steps by which protein complexes can assemble, using electrospray mass spectrometry experiments, literature-curated assembly data, and a large-scale analysis of protein complex structures. We found that most assembly steps can be classified into three basic types: dimerization, cyclization, and heteromeric subunit addition. By systematically combining different assembly steps in different ways, we were able to enumerate a large set of possible quaternary structure topologies, or patterns of key interfaces between the proteins within a complex. The vast majority of real protein complex structures lie within these topologies. This enables a natural organization of protein complexes into a “periodic table,” because each heteromer can be related to a simpler symmetric homomer topology. Exceptions are mostly the result of quaternary structure assignment errors, or cases where sequence-identical subunits can have different interactions and thus introduce asymmetry. Many of these asymmetric complexes fit the paradigm of a periodic table when their assembly role is considered. Finally, we implemented a model based on the periodic table, which predicts the expected frequencies of each quaternary structure topology, including those not yet observed. Our model correctly predicts quaternary structure topologies of recent crystal and electron microscopy structures that are not included in our original data set.
CONCLUSION: This work explains much of the observed distribution of known protein complexes in quaternary structure space and provides a framework for understanding their evolution. In addition, it can contribute considerably to the prediction and modeling of quaternary structures by specifying which topologies are most likely to be adopted by a complex with a given stoichiometry, potentially providing constraints for multi-subunit docking and hybrid methods. Lastly, it could help in the bioengineering of protein complexes by identifying which topologies are most likely to be stable, and thus which types of essential interfaces need to be engineered.