Portable devices provide rapid, accurate diagnosis of tuberculosis, other bacterial infections
May 7, 2013
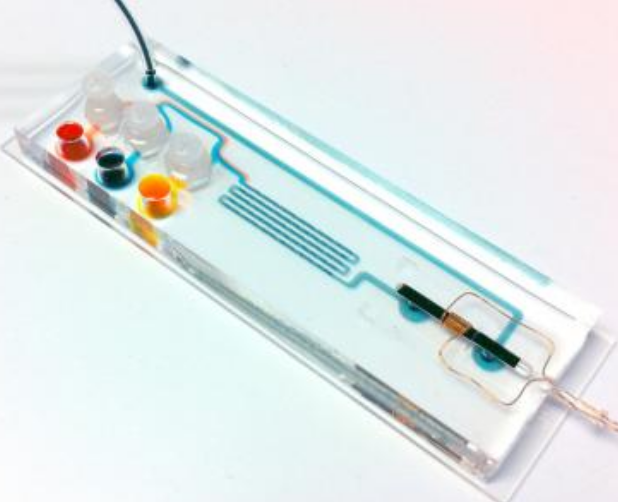
On this 2.5- by 7.5-cm cartridge, DNA extracted from sputum samples is amplified in the chambers on the left. TB-specific sequences are magnetically labeled in the microfluidic mixing channels in the center and detected by passage through the micro-NMR coil on the right. (Credit: Center for Systems Biology, Massachusetts General Hospital)
Two new portable diagnostic devices for rapid, accurate diagnosis of tuberculosis and other bacterial infections have been developed by researchers at Massachusetts General Hospital (MGH), Harvard Medical School, Harvard School of Public Health, and the Broad Institute.
A microfluidic device for diagnosing TB, other infectious bacteria
A handheld diagnostic device that MGH investigators first developed to diagnose cancer has been adapted to rapidly diagnose tuberculosis (TB) and other important infectious bacteria.
The portable device — about the size of a standard laboratory slide — combines microfluidic technology with nuclear magnetic resonance (NMR) to diagnose these important infections, and also to determine the presence of antibiotic-resistant bacterial strains.
“Rapidly identifying the pathogen responsible for an infection and testing for the presence of resistance are critical not only for diagnosis but also for deciding which antibiotics to give a patient,” says Ralph Weissleder, MD, PhD, director of the MGH Center for Systems Biology (CSB), a professor of Radiology at Harvard Medical School, and co-senior author of two papers in Nature Communications and Nature Nanotechnology.
“These described methods allow us to do this in two to three hours — a vast improvement over standard culturing practice, which can take as much as two weeks to provide a diagnosis.”
Investigators at the MGH CSB previously developed portable devices capable of detecting cancer biomarkers in the blood or in very small tissue samples. Target cells or molecules are first labeled with magnetic nanoparticles, and the sample is then passed through a micro NMR system capable of detecting and quantifying levels of the target.
But initial efforts to adapt the system to bacterial diagnosis had trouble finding antibodies — the detection method used in the earlier studies — that would accurately detect the specific bacteria. So the team switched to targeting specific nucleic acid sequences instead.
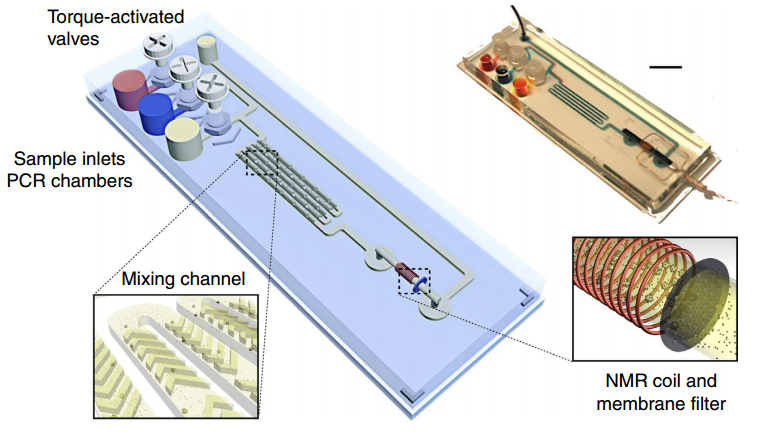
Magnetic barcode assay for sensitive TB detection. Whole-genomic DNA extracted from expectorated samples, capture beads, and magnetic nanoparticlse are loaded into inlet chambers gated by screw valves. After on-chip PCR, magnetic labelling of the beads takes place along the mixing channel. The magnetically barcoded beads are then purified and concentrated into the mNMR probe (microcoil) by the membrane filter. Scale bar, 1 cm. (Credit: Monty Liong et al./Nature Communications)
The new system detects DNA from the tuberculosis bacteria in small sputum samples. After DNA is extracted from the sample, any of the target sequence that is present is amplified using a standard procedure, then captured by polymer beads containing complementary nucleic acid sequences and labeled with magnetic nanoparticles with sequences that bind to other portions of the target DNA. The miniature NMR coil incorporated into the device detects any TB bacterial DNA present in the sample.
Tests of the device on samples from patients known to have TB and from healthy controls identified all positive samples, with no false positives, in less than three hours.Existing diagnostic procedures can take weeks to provide results and can miss up to 40 percent of infected patients.
Results were even stronger for patients infected with both TB and HIV — probably because infection with both pathogens leads to high levels of the TB bacteria — and specialized nucleic acid probes developed by the research team were able to distinguish treatment-resistant bacterial strains.
Hypersensitive ribosomal RNA system for detecting 13 important pathogens
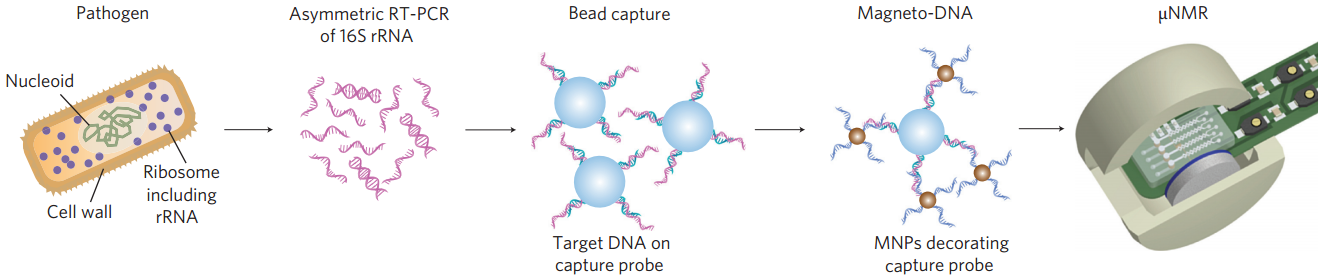
Schematic of magneto-DNA assay procedure for the detection of bacterial 16S rRNA. Total RNA is extracted from the specimen, and the 16S rRNA is amplified by asymmetric RT-PCR. Single-strand DNA of the amplified product is then captured by beads conjugated to capture probes, before hybridizing with magnetic nanoparticles to form a magnetic sandwich complex. Samples are subsequently analyzed using a microNMR system. (Credit: Hyun Jung Chung et al./Nature Nanotechnology)
The investigators also developed a similar system that uses ribrosomal RNA (rRNA) — already in use as a bacterial biomarker — as a target for nanoparticle labeling. The investigators developed both a universal nucleic acid probe that detects an rRNA region common to many bacterial species and a set of probes that target sequences specific to 13 clinically important pathogens, including Streptococcus pneumoniae, Escherichia coli and methicillin-resistant Staphylococcus aureus (MRSA).
The device was sensitive enough to detect as few as one or two bacteria in a 10 ml blood sample and to accurately estimate bacterial load. Testing the system on blood samples from patients with known infections accurately identified the particular bacterial species in less than two hours and also detected two species that had not been identified with standard culture techniques.
Ideal for developing countries
While both systems require further development to incorporate all steps into sealed, stand-alone devices, reducing the risk of contamination, Weissleder notes that the small size and ease of use of these devices make them ideal for use in developing countries.
“The magnetic interactions that pathogen detection is based on are very reliable, regardless of the quality of the sample, meaning that extensive purification (which would be difficult in resource-limited setting) is not necessary. The ability to diagnose TB in a matter of hours could allow testing and treatment decisions within the same clinic visit, which can be crucial to controlling the spread of TB in developing countries.”
Hakho Lee, PhD, of the MGH Center for Systems Biology, an assistant professor of Radiology at Harvard Medical School and co-senior author of both papers, notes that the system will also have important applications in developed countries.
“The capacity of the system not only to identify bacterial species but also to differentiate factors such as antibiotic resistance will help clinicians treat patients with the ‘right’ drugs from the start, which also helps reduce the emergence of treatment-resistant strains. The fact that this device requires only a tiny drop of the sample to be tested will be helpful in instances when specimens can be hard to obtain, such as treating children or seniors.”
Support for both studies includes National Institutes of Health grants.