Programming RNA to selectively kill mutated cells
September 9, 2010
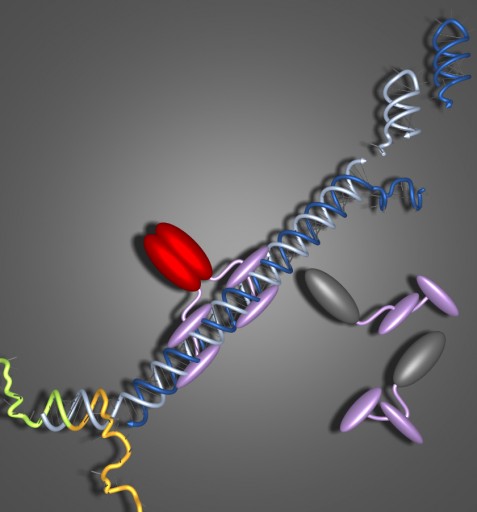
Small conditional RNAs selectively kill cancer cells. In lab-grown human brain, prostate and bone cancer cells, small conditional RNAs (light and dark blue) bind to a targeted RNA cancer mutation (orange and green), triggering self-assembly of a long double-stranded RNA polymer that activates an innate immune response (gray turns to red) leading to cell death. No measurable reduction in numbers is observed for cells lacking targeted cancer mutations. (Suvir Venkataraman, William M. Clemons, Jr. and Niles A. Piercel / Caltech)
What if we had cancer treatments that worked more like a computer program, which can perform actions based on conditional statements? Then, a treatment would kill a cell if–and only if–the cell had been diagnosed with a mutation. Only the defective cells would be destroyed, virtually eliminating unwanted side effects.
With support from the National Science Foundation (NSF), researchers at the California Institute of Technology have created conditional small RNA molecules to perform this task. Their strategy uses characteristics that are built into our DNA and RNA to separate the diagnosis and treatment steps.
“The molecules are able to detect a mutation within a cancer cell, and then change conformation to activate a therapeutic response in the cancer cell, while remaining inactive in cells that lack the cancer mutation,” claims Niles Pierce, co-author of a recent study which appears in the September 6 issue of Proceedings of the National Academy of Sciences (PNAS).
This work is part of the Molecular Programming Project, funded by NSF’s Directorate for Computer & Information Science & Engineering. One of the goals of the project is to increase understanding of how information can be stored and processed by molecules, and how we might create practical applications that utilize that information.
RNAs perform all kinds of functions in a cell, acting as messengers and switches to communicate and monitor which genes are expressed in a cell at any given time. A particular class of RNAs, called small RNAs, is less than 30 base pairs in length (an average gene is thousands of base pairs long). These small bits of RNA are involved in many of the processes that maintain life. The treatment developed by Pierce and his colleagues relies on two separate small RNAs that structurally mimic those that occur naturally within our own cells. Because these molecules resemble small RNAs that are normally present, the researchers hope there will be few, if any side effects.
“By decoupling diagnosis and treatment, we can create molecules that are both highly selective and highly effective in killing cancer cells,” said Pierce. “Conceptually, small conditional RNAs have the potential to transform cancer treatment because they change what we can expect from a molecule. Many years of work remain to establish whether this conceptual promise can be realized in human patients.”
How it works
Treatment involves two different small RNAS. The first small RNA will open up if–and only if–it finds the cancer mutation. A positive “diagnosis” exposes a signal that was previously hidden within the small RNA. Once this small RNA is open, a second small RNA binds to it, setting off a chain reaction in which these RNA molecules continue to combine to form a longer chain. The length of the chain is an important part of the “treatment.” Longer chains trick the cell into thinking it has been invaded by a virus, tripping a self-destruct response.
In the PNAS study, researchers demonstrated that this approach effectively eliminates lab-grown human brain, prostate and bone cancer cells in a mutation-specific manner. Future experiments will determine whether the treatment is effective on a larger scale.
More info: National Science Foundation news