Sequenced microbial genomes could help increase our resistance to disease
June 11, 2010
The first 178 reference microbial genomes have now been analyzed and catalogued by the he Human Microbiome Project (HMP) by researchers with the U.S. Department of Energy’s Lawrence Berkeley National Laboratory.
A microbiome is the full complement of microorganisms populating a “supraorganism,” such as a human. These symbiotic microbes outnumber an adult’s own somatic and germ cells by up to a 10:1 ratio; the interactions between our human and microbial cells go a long way towards determining your health and physical well-being, especially our resistance to infectious diseases.
The goal of the HMP is to sequence the genomes of 1,000 or more of these microbial species and assemble the information in a “project catalog” as a reference for future investigations.
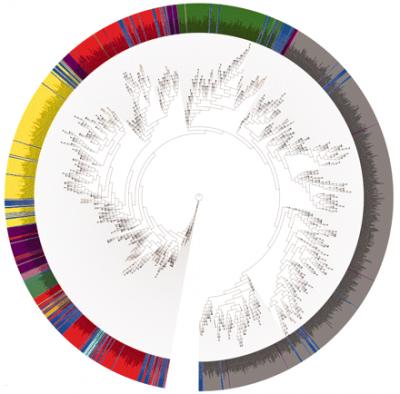
Phylogenetic analysis of 16S Ribosomal DNA sequences with Human Microbiome Project microbes highlighted in blue shows the distribution of these human symbiants around the microbial tree of life. Phylum are separated by color as follows: yellow, Actinobacteria; dark green, Bacteroidetes; light green, Cyanobacteria; red, Firmicutes; cyan, Fusobacteria; dark red, Planctomycetes; gray, Proteobacteria; magenta, Spirochaetes; light pink, TM7; tan, Tenericutes. (Human Microbiome Project)
More info: DOE/Lawrence Berkeley National Laboratory