Text mining: what do publishers have against this high-tech research tool?
May 25, 2012 | Source: The Guardian
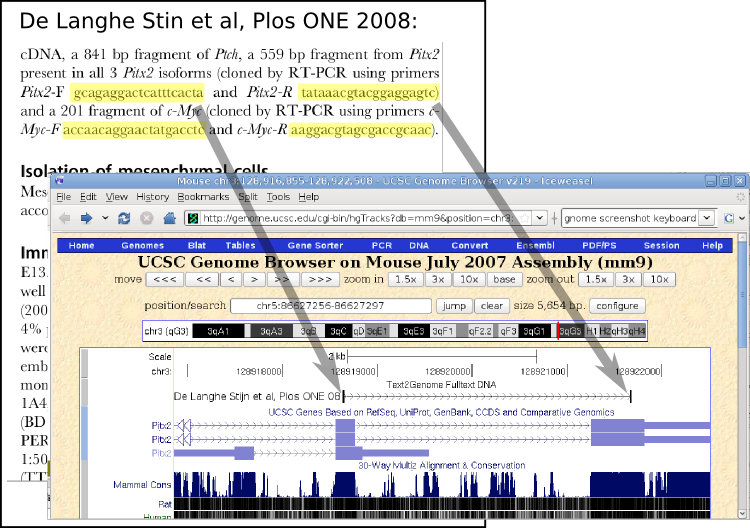
text2Genome is using a unique way to map scientific articles to genomic locations: From a full-text scientific article and it's supplementary data files, all words that resemble DNA sequences are extracted and then mapped to public genome sequences. They can then be displayed on genome browser websites and used in data-mining applications. (Credit: text2Genome)
Researchers are pushing to make tens of thousands of papers based on publicly funded research work available through open access to find links between genes and diseases — countering publishers’ default ban on text mining via computer scanning.
That would allow researchers to mine the content freely without needing to request any extra permissions.
Researchers often need access to tens of thousands of research papers at once, so they can use computers to look for unseen patterns and associations across the millions of words in the articles.
This technique, called text mining, is a vital 21st-century research method. It uses powerful computers to find links between drugs and side effects, or genes and diseases, that are hidden within the vast scientific literature. These are discoveries that a person scouring through papers one by one may never notice.
It is a technique with big potential. A report published by McKinsey Global Institute last year said that “big data” technologies such as text and data mining had the potential to create €250bn (£200bn) of annual value to Europe’s economy, if researchers were allowed to make full use of it.
The scale of new information in modern science is staggering: more than 1.5 million scholarly articles are published every year and the volume of data doubles every three years. No individual can keep up with such a volume, and scientists need computers to help them digest and make sense of the information.