Victory for crowdsourced biomolecule design
January 23, 2012 | Source: Nature News
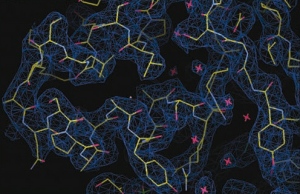
An enzyme designed by players of the protein-folding game Foldit was better than anything scientists could come up with (credit: Eiben, C. B. et al.)
Obsessive gamers’ hours at the computer have now topped scientists’ efforts to improve a model enzyme, in the first crowdsourced redesign of a protein.
The online game Foldit allows players to fiddle at folding proteins on their home computers in search of the best-scoring (lowest-energy) configurations.
By posing a series of puzzles to Foldit players and then testing variations on the players’ best designs in the lab, researchers have created an enzyme with more than 18-fold higher activity than the original.
There are no immediate applications for the particular reaction that this enzyme catalyzes,but it marks a milestone in showing what crowdsourcing research can achieve.
The researchers are now looking toward more useful targets. The team reported last year that they had designed small protein inhibitors that bind to and block the 1918 pandemic influenza virus. Foldit players are working to make more potent inhibitors, which could be drugs.
Ref.: Eiben, C. B. et al., Increased Diels-Alderase activity through backbone remodeling guided by Foldit players, Nature Biotechnology, 2012 [DOI: 10.1038/nbt.2109]
Ref.: Eiben, C. B. et al., Increased Diels-Alderase activity through backbone remodeling guided by Foldit players, Nature Biotechnology, 2012 [PDF]