Viruses act like ‘self-packing suitcases’
October 22, 2012
Researchers at the University of Leeds have identified a crucial stage in the life cycle of simple viruses like polio and the common cold that could open a new front in the war on viral disease.
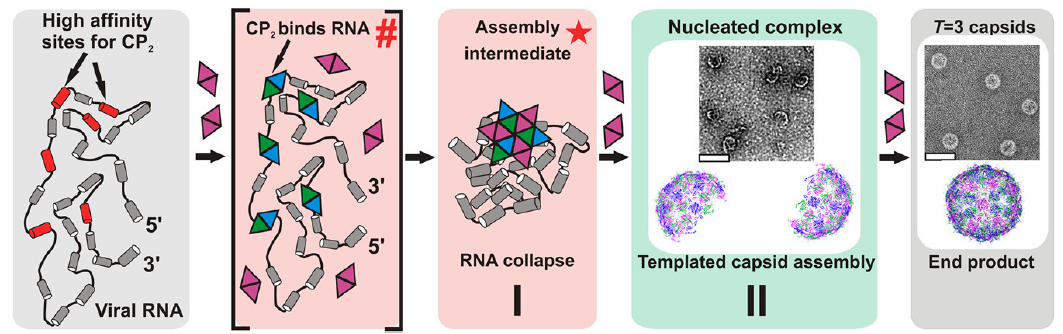
The team is the first to observe at a single-molecule level how the genetic material (genome) that forms the core of a single-strand RNA virus particle packs itself into its outer shell of proteins — overturning accepted thinking about the process and opening a chink in the armor of a wide range of viruses.
“If we can target this process, it could lead to a completely new class of anti-virals that would be less likely to create resistant viruses than existing drugs, which tend to target individual proteins,” Professor Peter Stockley said.
A number of important viruses like the common cold and polio have RNA (ribonucleic acid) instead of DNA as their genetic material. The observations reveal that the viruses’ RNA initially has a much greater volume than the virus particles created after they are packed inside their protein shell. “We realized that the RNA genome must have to be intricately folded to fit into the final container,” said co-author Dr. Roman Tuma from the University of Leeds’ Faculty of Biological Sciences.
How to open the ‘suitcase’
When the team added proteins to the viral RNA they saw an immediate collapse in its volume. “It seems that viral RNAs have evolved a self-folding mechanism that makes closing the ‘viral suitcase’ very efficient. It’s as though ‘the suitcase and the ‘clothes’ work together to close the lid and protect the content,” Tuma said.
“The viral RNAs, and only the viral RNAs, can do this trick of folding up to fit as soon as they see the ‘suitcase’ coming. That’s the important thing. If we can interfere in that process, we’ve got a completely novel drug target in the lifecycle of viruses,” Professor Stockley said.
“At the moment there are relatively few antiviral drugs and they tend to target enzymes that the virus encodes in its genome. The problem is that the drugs target one enzyme initially and, within the year, scientists are identifying strains that have become resistant. Individual proteins are extremely susceptible to this mutation. A fundamental process like the one we’re looking at opens the possibility of targeting the collective behavior of essential molecules, which could be much less susceptible to developing resistance,” explained Professor Stockley.
The same phenomenon is seen in both bacterial and plant viruses. “While we have not proved it yet, I would put money on animal viruses showing the same mechanism too,” Professor Stockley added.
The team used sophisticated instrumentation custom built at the University that allowed them to make the first ever single-molecule measurements of viral assembly. This allowed researchers to observe individual viral particles one at a time. “The specific collapse, which can only be seen in such assays, was totally unexpected and overturns the current thinking about assembly,” Professor Stockley said.