Zeroing in on how Alzheimer’s-disease toxins are created
February 19, 2014
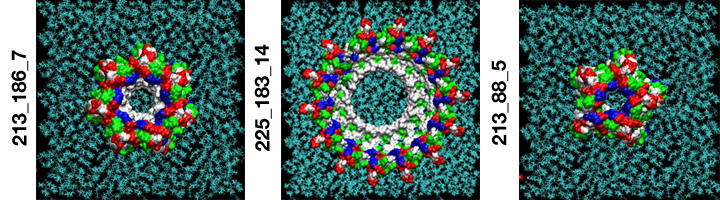
Single amyloid-beta monomers can pair up to form a variety of dimers that can aggregate into larger peptide rings that reside on cell membranes such as those pictured. This process has been implicated in the development of Alzheimer’s disease. This visualization shows the possible rings which have the most favorable energies of interactions with the membrane. The residues are colored white to represent apolar or hydrophobic areas, green for the polar or hydrophilic areas, blue to show a positive charge, and red to show a negative charge. (Credit: Igor Tsigelny, SDSC and UC San Diego; Eliezer Masliah, UC San Diego)
Using the Gordon supercomputer at the San Diego Supercomputer Center (SDSC) at the University of California, San Diego, researchers have achieved new insights into how toxic molecular complexes associated with Alzheimer’s disease are created.
Igor Tsigelny, a research scientist with SDSC, the UCSD Moores Cancer Center, and the Department of Neurosciences, focused on a small peptide called amyloid-beta, which pairs up with itself to form dimers and oligomers — different forms of molecular complexes.
The scientists surveyed all the possible ways to look at the dynamics of conformational changes of these peptides and the possibility that they might organize into the oligomers theorized to be responsible for the degenerative brain disease. In the February 14 issue of the Journal of Alzheimer’s Disease (open access), the researchers suggest their results may generate new targets for drug development.
“Our research has identified amino acids for point mutations that either enhanced or suppressed the formation and toxicity of oligomer rings,” said Tsigelny, the study’s lead author. “Aggregation of misfolded neuronal proteins and peptides may play a primary role in neurodegenerative disorders, including Alzheimer’s disease.”
Key role of amyloid-beta dimers
Tsigelny also noted that recent improvements in computational processing speed have allowed him and other researchers to use a variety of tools, including computer simulations, to take new approaches to examining amyloid-beta, which has proven too unstable for traditional approaches such as x-ray crystallography.
According to the researchers, their work implicates a more dynamic role for amyloid-beta dimers than previously thought. It also suggests that the way dimers form and then grow into larger structures is a rapidly changing process.
“This, as well as previous results, suggests that targeting selected amyloid-beta dimers may be important in an effort to ameliorate the episodic memory described in mild cognitive impairment and the early stages of Alzheimer’s disease,” said Eliezer Masliah, professor of pathology and medicine at UC San Diego, and a member of the research team.
Abstract of Journal of Alzheimer’s Disease paper
Alzheimer’s disease (AD) is associated with the formation of toxic amyloid-β (Aβ)42 oligomers, and recent evidence supports a role for Aβ dimers as building blocks for oligomers. Molecular dynamics simulation studies have identified clans for the dominant conformations of Aβ42 forming dimers; however, it is unclear if a larger spectrum of dimers is involved and which set(s) of dimers might evolve to oligomers verse fibrils. Therefore, for this study we generated multiple structural conformations of Aβ42, using explicit all-atom molecular dynamics, and then clustering the different structures based on key conformational similarities. Those matching a selection threshold were then used to model a process of oligomerization. Remarkably, we showed a greater diversity in Aβ dimers than previously described. Depending on the clan family, different types of Aβ dimers were obtained. While some had the tendency to evolve into oligomeric rings, others formed fibrils of diverse characteristics. Then we selected the dimers that would evolve to membranephilic annular oligomers. Nearly one third of the 28 evaluated annular oligomers had the dimer interfaces between the neighboring Aβ42 monomers with possible salt bridges between the residue K28 from one side and either residue E22 or D23 on the other. Based on these results, key amino acids were identified for point mutations that either enhanced or suppressed the formation and toxicity of oligomer rings. Our studies suggest a greater diversity of Aβ dimers. Understanding the structure of Aβ dimers might be important for the rationale design of small molecules that block formation of toxic oligomers.