Capturing ultrasharp images of multiple cell components simultaneously
February 12, 2014
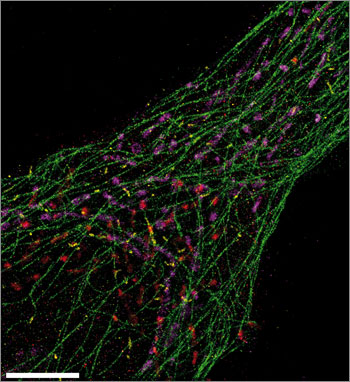
This ultrasharp Exchange-PAINT image simultaneously spots microtubules (green), mitochondria (purple), Golgi apparatus (red), and peroxisomes (yellow) from a single human cell. Scale bar: 5 µm. (Credit: Maier Avendano/Wyss Institute)
A new microscopy method could enable scientists to generate images of dozens of different biomolecules in a human cell simultaneously, a team from the Wyss Institute of Biologically Inspired Engineering at Harvard University reported in Nature Methods.
Such images could shed light on complex cellular pathways and potentially lead to new ways to diagnose disease, track its prognosis, or monitor the effectiveness of therapies at a cellular level.
Cells often employ dozens or even hundreds of different proteins and RNA molecules to get a complex job done. Today’s methods typically only spot at most three or four types of these tiny workers simultaneously.
To understand complex cellular functions, it’s important to be able to visualize most or all of those workers at once, said Peng Yin, Ph.D., a Core Faculty member at the Wyss Institute and Assistant Professor of Systems Biology at Harvard Medical School.
Problem: when two objects are closer than about 200 nanometers apart they cannot be distinguished using a traditional light microscope: the viewer sees one blurry blob where in reality there are two objects. This is known as the Abbe diffraction limit.
Since the mid-1990s, scientists have developed several “super-resolution” methods to overcome this problem, using combinations of specialized optics, special fluorescent proteins, or dyes that tag cellular components.
Super-resolution
Ralf Jungmann, Ph.D., now a Postdoctoral Fellow working with Yin at the Wyss Institute and Harvard Medical School, helped develop one of those super-resolution methods, called DNA-PAINT, as a graduate student. DNA-PAINT can create ultrasharp snapshots of up to three cellular workers at once by labeling them with different colored dyes.
To visualize cellular job sites with crews of dozens of cellular workers, Yin’s team modified DNA-PAINT to create a new method called Exchange-PAINT.
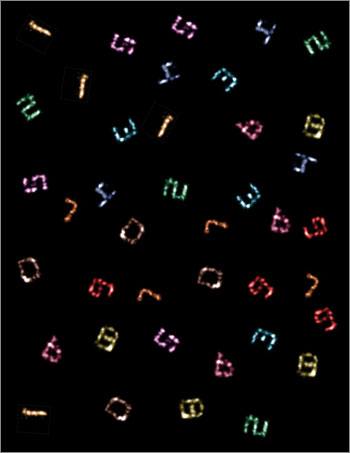
Ten-color super-resolution images (artificially arranged into one composite) of synthetic DNA nanostructures displaying patterns that resemble the numerals “0” through “9,” produced using only a single dye and a single laser source (credit: Johannes B. Woehrstein/Wyss Institute)
Exchange-PAINT relies on the fact that DNA strands with the correct sequence of letters, or nucleotides, bind specifically to partner strands with complementary sequences.
The researchers label a biomolecule they want to visualize with a short DNA tag, then add to the solution a partner strand carrying a fluorescent dye that lights up only when the two strands pair up.
When that partner strand binds the tagged biomolecule, it lights up, then lets go, causing the biomolecule to “blink” at a precise rate the researchers can control. The researchers use this blinking to obtain ultrasharp images.
They then repeat the process to visualize a second target, a third, and so on. Then they overlay the resulting images to create a composite image in which each biomolecule — each cellular worker — is assigned a different color.
This allows them to create false-color images that simultaneously show many types of biomolecules — far more than they could simultaneously visualize by labeling them with different colored dyes. And these false-color images allow them to spot enough cellular workers at once to capture the entire scene.
Testing with DNA origami
To test Exchange-PAINT, the researchers created 10 unique pieces of folded DNA, or DNA origami, that resembled the numerals 0 through 9. These numerals could be resolved at less than 10 nanometers resolution, or one-twentieth of the diffraction limit.
The team was able to use Exchange-PAINT to capture clear images of the 10 different types of miniscule DNA origami structures in one image. They also used the method to capture detailed, ultrasharp images of fixed human cells, with each color tagging an important cellular component — microtubules, mitochondria, Golgi apparatus, or peroxisomes.
Yin expects the method, with further development, to be able to visualize dozens of cellular components at once.
The work was funded by the National Institutes of Health, the Office of Naval Research, the National Science Foundation, the Humboldt Foundation, and the Wyss Institute.
Abstract of Nature Methods paper
Super-resolution fluorescence microscopy is a powerful tool for biological research, but obtaining multiplexed images for a large number of distinct target species remains challenging. Here we use the transient binding of short fluorescently labeled oligonucleotides (DNA-PAINT, a variation of point accumulation for imaging in nanoscale topography) for simple and easy-to-implement multiplexed super-resolution imaging that achieves sub-10-nm spatial resolution in vitro on synthetic DNA structures. We also report a multiplexing approach (Exchange-PAINT) that allows sequential imaging of multiple targets using only a single dye and a single laser source. We experimentally demonstrate ten-color super-resolution imaging in vitro on synthetic DNA structures as well as four-color two-dimensional (2D) imaging and three-color 3D imaging of proteins in fixed cells.