Chips that listen to bacteria
February 18, 2014
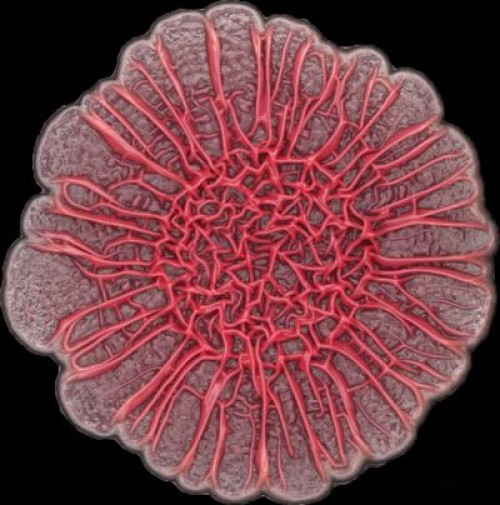
The development of colony biofilms by Pseudomonas aeruginosa is affected by redox-active compounds called phenazines (credit: Hassan Sakhtah, Columbia University)
A Columbia University research team has demonstrated that integrated circuit technology can be used for a study of signaling in bacterial colonies.
The researchers have developed a complementary metal-oxide-semiconductor (CMOS) chip that enables them to electrochemically image the signaling molecules from these colonies spatially and temporally.
In effect, they have developed chips that “listen” to bacteria.
“This is an exciting new application for CMOS technology that will provide new insights into how biofilms form,” says Ken Shepard, professor of electrical engineering and biomedical engineering at Columbia Engineering.
“Disrupting biofilm formation has important implications in public health in reducing infection rates.”
Detecting electrons instead of photons
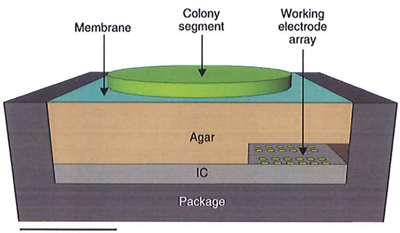
Experimental electrosensor setup, including the bacterial colony, integrated circuit, and agar interface membrane. Scale bar: 1 mm. (Credit: Daniel L. Bellin et al./Nature Communications)
The chip, which senses the electron charge on molecules, is capable of imaging 60 pixels for concentrations as low as 2.6 microns (millionths of a meter) in 36 seconds.
The researchers say that this is the first time integrated circuits have been used for imaging small molecules electrochemically in a multicellular structure. Optical microscopy techniques cannot directly detect critical components of physiology, such as primary metabolism and signaling factors.
Cells mediate their physiological activities using secreted molecules, explains Lars Dietrich, assistant professor of biological sciences at Columbia University.
The team looked specifically at phenazines, which are secreted metabolites that control gene expression. Their study found that the bacterial colonies produced a phenazine gradient that, they say, is likely to be of physiological significance and contribute to colony morphogenesis.
Disrupting biofilms in medical devices
“This is a big step forward,” Dietrich continues. “We describe using this chip to ‘listen in’ on conversations taking place in biofilms, but we are also proposing to use it to interrupt these conversations and thereby disrupt the biofilm (a special concern in hospitals). In addition to the pure science implications of these studies, a potential application of this would be to integrate such chips into medical devices that are common sites of biofilm formation, such as catheters, and then use the chips to limit bacterial colonization.”
The next step for the team will be to develop a larger chip that will enable larger colonies to be imaged at higher spatial and temporal resolutions.
“This represents a new and exciting way in which solid-state electronics can be used to study biological systems,” Shepard adds. “This is one of the many emerging ways integrated circuit technology is having impact in biotechnology and the life sciences.”
The study, published in Nature Communications, was supported by the National Institutes of Health and the National Science Foundation.
Abstract of Nature Communications paper
Despite advances in monitoring spatiotemporal expression patterns of genes and proteins with fluorescent probes, direct detection of metabolites and small molecules remains challenging. A technique for spatially resolved detection of small molecules would benefit the study of redox-active metabolites that are produced by microbial biofilms and can affect their development. Here we present an integrated circuit-based electrochemical sensing platform featuring an array of working electrodes and parallel potentiostat channels. ‘Images’ over a 3.25 × 0.9 mm2 area can be captured with a diffusion-limited spatial resolution of 750 μm. We demonstrate that square wave voltammetry can be used to detect, identify and quantify (for concentrations as low as 2.6 μM) four distinct redox-active metabolites called phenazines. We characterize phenazine production in both wild-type and mutant Pseudomonas aeruginosa PA14 colony biofilms, and find correlations with fluorescent reporter imaging of phenazine biosynthetic gene expression.