CRISPR enhancements: improving the ability to delete genes
March 9, 2016
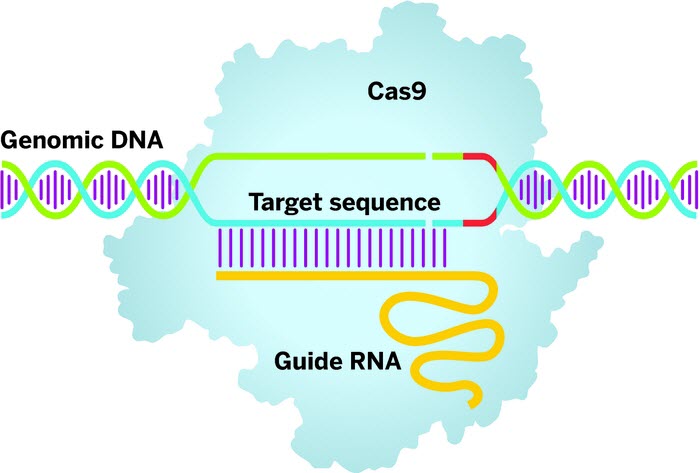
Clustered regularly interspaced short palindromic repeats (CRISPRs) technology employs a guide RNA to direct the Cas9 enzyme (light blue) to a target DNA sequence. Once there, Cas9 will bind when it finds a protospacer-adjacent motif sequence (red) in the DNA and cut both strands, priming the gene sequence for editing. (credit: Adapted from OriGene Technologies)
Scientists have found a way to improve the efficiency of the controversial gene editing technology, CRISPR/Cas9 (“CRISPR”).
Lauded as a groundbreaking technology that allows scientists to modify genes* for many different applications, CRISPR/Cas9 has hit stormy waters over the ethics of editing human embryos. Although the technology is faster and cheaper than past gene editing techniques, one of the problems cited is that the efficiency of deleting unwanted genes is low and that it gives inconsistent results — an unacceptable risk when using human embryos.
Haoquan Wu, PhD, from the Texas Tech University Health Sciences Center El Paso, has worked to improve CRISPR’s overall ability to target and eliminate genes.
In an open-access paper published in Genome Biology, senior author Wu and his team describe how they were able to significantly improve the efficiency of gene knockout (deletion) by creating structural changes to the CRISPR RNA guide molecule.**
“The extent of the improvement in knockout efficiency with these changes was striking,” Wu says. “This is going to help reduce concerns that knockout experiments might not work, and also significantly increase the efficiency of more challenging editing procedures like gene deletion.”
At this stage, the researchers are unsure why the changes to the single guide RNA enhanced CRISPR’s efficiency, although options include an enhanced ability to bind with Cas9 or improved stability of the RNA.
The TTUHSC El Paso team plans to continue studying this modified single guide RNA template to better understand why it enhances CRISPR’s functionality. They’ve also applied for a patent for their new method, which they hope will be adopted by other scientists using CRISPR.
* CRISPR/Cas9 has two main components. A small “single guide” RNA molecule (Clustered Regularly Interspaced Short Palindromic Repeats, CRISPR) directs the Cas9 enzyme to a specific sequence of DNA. The Cas9 then acts like molecular scissors to cut that specific piece of DNA. The DNA is then ready for editing.
** Some technical aspects of CRSIPR-Cas9 are inefficient, including total gene knockout. To improve the efficiency, the researchers optimized the structure of the small-guide RNA by tweaking the sequence and making it slightly longer. The optimized small-guide RNA was far more efficient at producing knockouts — 18 of 24 experiments produced greater than 50 per cent knockout efficiency compared with only four for the original small-guide RNA structure.
Abstract of Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency
Background: Single-guide RNA (sgRNA) is one of the two key components of the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 genome-editing system. The current commonly used sgRNA structure has a shortened duplex compared with the native bacterial CRISPR RNA (crRNA)–transactivating crRNA (tracrRNA) duplex and contains a continuous sequence of thymines, which is the pause signal for RNA polymerase III and thus could potentially reduce transcription efficiency.
Results: Here, we systematically investigate the effect of these two elements on knockout efficiency and showed that modifying the sgRNA structure by extending the duplex length and mutating the fourth thymine of the continuous sequence of thymines to cytosine or guanine significantly, and sometimes dramatically, improves knockout efficiency in cells. In addition, the optimized sgRNA structure also significantly increases the efficiency of more challenging genome-editing procedures, such as gene deletion, which is important for inducing a loss of function in non-coding genes.
Conclusions: By a systematic investigation of sgRNA structure we find that extending the duplex by approximately 5 bp combined with mutating the continuous sequence of thymines at position 4 to cytosine or guanine significantly increases gene knockout efficiency in CRISPR-Cas9-based genome editing experiments.