Discovering new drugs and materials by ‘touching’ molecules in virtual reality
July 6, 2018
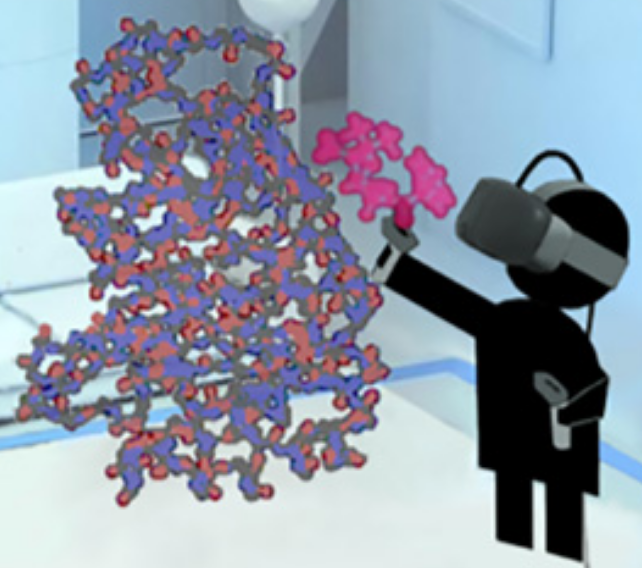
To figure out how to block a bacteria’s attempt to create multi-resistance to antibiotics, a researcher grabs a simulated ligand (binding molecule) — a type of penicillin called benzylpenicillin (red) — and interactively guides that molecule to dock within a larger enzyme molecule (blue-orange) called β-lactamase, which is produced by bacteria in an attempt to disable penicillin (making a patient resistant to a class of antibiotics called β-lactam). (credit: University of Bristol)
University of Bristol researchers, in collaboration with developers at Bristol based start-up Interactive Scientific, Oracle Corporation and a joint team of computer science and chemistry researchers, have designed and tested a new virtual reality (VR) cloud-based system intended to allow researchers to reach out and “touch” molecules as they move — folding them, knotting them, plucking them, and changing their shape to test how the molecules interact. The virtual reality cloud based system, called Nano Simbox, is the proprietary technology of Interactive Scientific, who collaborated with the University of Bristol to do the testing. Using an HTC Vive virtual-reality device, it could lead to creating new drugs and materials and improving the teaching of chemistry.
More broadly, the goal is to accelerate progress in nanoscale molecular engineering areas that include conformational mapping, drug development, synthetic biology, and catalyst design.
Real-time collaboration via the cloud

Two users passing a fullerene (C60) molecule back and forth in real time over a cloud-based network. The researchers are each wearing a VR head-mounted display (HMD) and holding two small wireless controllers that function as atomic “tweezers” to manipulate the real-time molecular dynamic of the C60 molecule. Each user’s position is determined using a real-time optical tracking system composed of synchronized infrared light sources, running locally on a GPU-accelerated computer. (credit: University of Bristol)
The multi-user system, developed by developed by a team led by University of Bristol chemists and computer scientists, uses an “interactive molecular dynamics virtual reality” (iMD VR) app that allows users to visualize and sample (with atomic-level precision) the structures and dynamics of complex molecular structures “on the fly” and to interact with other users in the same virtual environment.
Because each VR client has access to global position data of all other users, any user can see through his/her headset a co-located visual representation of all other users at the same time. So far, the system has uniquely allowed for simultaneously co-locating six users in the same room within the same simulation.
Testing on challenging molecular tasks
The team designed a series of molecular tasks for testing, using traditional mouse, keyboard, and touchscreens compared to virtual reality. The tasks included threading a small molecule through a nanotube, changing the screw-sense of a small organic helix, and tying a small string-like protein into a simple knot, and a variety of dynamic molecular problems, such as binding drugs to its target, protein folding, and chemical reactions. The researchers found that for complex 3D tasks, VR offers a significant advantage over current methods. For example, participants were ten times more likely to succeed in difficult tasks such as molecular knot tying.
Anyone can try out the tasks described in the open-access paper by downloading the software and launching their own cloud-hosted session.
David Glowacki | This video, made by University of Bristol PhD student Helen M. Deeks, shows the actions she took using a wireless set of “atomic tweezers” (using the HTC Vive) to interactively dock a single benzylpenicillin drug molecule into the active site of the β-lactamase enzyme.
David Glowacki | The video shows the cloud-mounted virtual reality framework, with several different views overlaid to give a sense of how the interaction works. The video outlines the four different parts of the user studies: (1) manipulation of buckminsterfullerene, enabling users to familarize themselves with the interactive controls; (2) threading a methane molecule through a nanotube; (3) changing the screw-sense of a helicene molecule; and (4) tying a trefoil knot in 17-Alanine.
Ref: Science Advances (open-access). Source: University of Bristol.