DNA-guided 3-D printing of human tissue
September 22, 2015
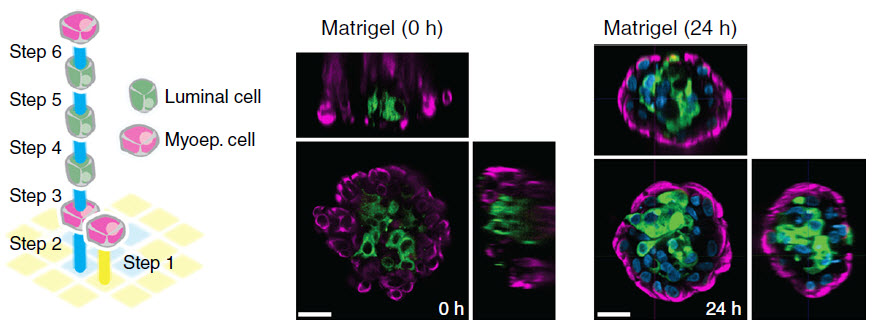
Reconstituting epithelial (skin) microtissues with programmed size, shape, composition, spatial heterogeneity, and embedding extracellular matrix. Scheme and images of fully embedded aggregates of human luminal and myoepithelial cells. (credit: Michael E Todhunter et al./Nature Methods)
A new technique developed by UCSF scientists for building organoids (tiny models of human tissues) more precisely turns human cells into the biological equivalent of LEGO bricks. Called DNA Programmed Assembly of Cells (DPAC), it allows researchers in hours to create arrays of thousands of custom-designed organoids, such as models of human mammary glands containing several hundred cells each.
These mini-tissues in a dish can be used to study how particular structural features of tissue affect normal growth or go awry in cancer. They could be used for therapeutic drug screening and to help teach researchers how to grow whole human organs.
The new technique, reported in an open-the journal Nature Methods on Aug. 31, allows for researchers to “take any cell type we want and program just where it goes,” said Zev Gartner, PhD, the paper’s senior author and an associate professor of pharmaceutical chemistry at UCSF. “We can precisely control who’s talking to whom and who’s touching whom at the earliest stages.”
There are very few limits to the tissues this technology can mimic, he said. “One potential application would be that within the next couple of years, we could be taking samples of different components of a cancer patient’s mammary gland and building a model of their tissue to use as a personalized drug screening platform. Another is to use the rules of tissue growth we learn with these models to one day grow complete organs.”
Studying how the cells of complex tissues like the mammary gland self-organize, make decisions as groups, and break down in disease has been a challenge to researchers. The living organism is often too complex to identify the specific causes of a particular cellular behavior. On the other hand, cells in a dish lack the critical element of realistic 3-D structure.
DNA as molecular Velcro and bar code
To specify the 3-D structure of their organoids, the researchers incubate cells with tiny snippets of single-stranded DNA engineered to slip into the cells’ outer membranes, covering each cell like the hairs on a tennis ball. These DNA strands act both as a sort of molecular Velcro and as a bar code that specifies where each cell belongs within the organoid. When two cells incubated with complementary DNA strands come in contact, they stick fast. If the DNA sequences don’t match, the cells float on by. Cells can be incubated with several sets of DNA bar codes to specify multiple allowable partners.
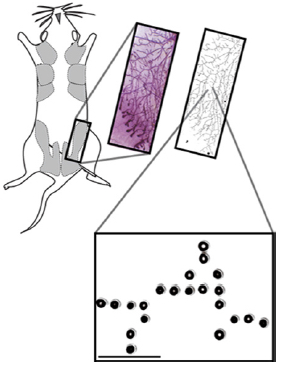
A whole-mount image of a digitized mouse mammary fat pad (reproduced with permission of W. Muller) used to print a pattern of DNA spots, and rendered as a 1.6-cm-long pattern of single cells fully embedded in gelatinous protein mixture (credit: Michael E Todhunter et al./Nature Methods)
To turn these cellular LEGOs into arrays of organoids that can be used for research, Gartner’s team lays down the cells in layers, with multiple sets of cells designed to stick to particular partners. This lets them build up complex tissue components like the mammary gland. It also lets them experiment with specifically adding in a single cell with a known cancer mutation to different parts of the organoid to observe its effects.
To demonstrate the precision of the technique and its ability to generalize to many different human tissue types, the research team created several proof-of-principle organoid arrays mimicking human tissues such as branching vasculature and mammary glands.
In one experiment, the researchers created arrays of mammary epithelial cells and asked how adding one or more cells expressing low levels of the cancer gene RasG12V affected the cells around them. They found that normal cells grow faster when in an organoid with cells expressing RasG12V at low levels, but required more than one mutant cell to kick-start this abnormal growth. They also found that placing cells with low RasG12V expression at the end of a tube of normal cells allowed the mutant cells to branch and grow, drawing normal cells behind them like a bud at the tip of a growing tree branch.
Gartner’s group plans to use the technique to investigate what cellular or structural changes in mammary glands can lead to the breakdown of tissue architecture associated with tumors that metastasize, invading other parts of the body and threatening the life of the patient. They also hope to use what they learn from simple models of different tissue types to ultimately build functional human tissues like lung and kidney and neural circuits using larger-scale techniques.
Abstract of Programmed synthesis of three-dimensional tissues
Reconstituting tissues from their cellular building blocks facilitates the modeling of morphogenesis, homeostasis and disease in vitro. Here we describe DNA-programmed assembly of cells (DPAC), a method to reconstitute the multicellular organization of organoid-like tissues having programmed size, shape, composition and spatial heterogeneity. DPAC uses dissociated cells that are chemically functionalized with degradable oligonucleotide ‘Velcro’, allowing rapid, specific and reversible cell adhesion to other surfaces coated with complementary DNA sequences. DNA-patterned substrates function as removable and adhesive templates, and layer-by-layer DNA-programmed assembly builds arrays of tissues into the third dimension above the template. DNase releases completed arrays of organoid-like microtissues from the template concomitant with full embedding in a variety of extracellular matrix (ECM) gels. DPAC positions subpopulations of cells with single-cell spatial resolution and generates cultures several centimeters long. We used DPAC to explore the impact of ECM composition, heterotypic cell-cell interactions and patterns of signaling heterogeneity on collective cell behaviors.