Folding (synthesizing) artificial proteins
August 24, 2012
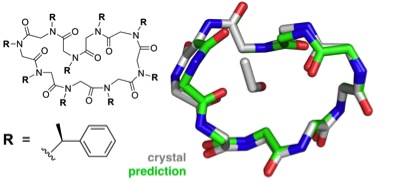
Predicted structure of the cyclic nonamer proposed by the theorists, shown to match the actual folded structure with remarkable accuracy (credit: Butterfoss et al./PNAS)
A computer modeling approach similar to one used to predict protein structures can also accurately predict peptoid (artificial protein) structure, a team of scientists has demonstrated.
Background
In the world of proteins, form defines function. Based on interactions between their constituent amino acids, proteins form specific conformations, folding and twisting into distinct, chemically directed shapes. The resulting structure dictates the proteins’ actions; thus accurate modeling of structure is vital to understanding functionality.
Peptoids, the synthetic cousins of proteins, follow similar design rules. Less vulnerable to chemical or metabolic breakdown than proteins, peptoids are promising for diagnostics, pharmaceuticals, and as a platform to build bioinspired nanomaterials, as scientists can build and manipulate peptoids with great precision.
But to design peptoids for a specific function, scientists need to first untangle the complex relationship between a peptoid’s composition and its function-defining folded structure.
Self-assembling artificial molecular machines
“Natural selection has engineered protein sequences that can self-assemble into molecular machines with specific functions. Why can’t we do the same with biologically inspired synthetic materials?” Vincent A.Voelz, Principal Investigator with Temple University, explains.
With this mission in mind, the collaborative team of scientists developed the project two years ago. “Some of the team [had] worked together on this truly difficult problem for almost 20 years,” says Kent Kirshenbaum of NYU.
They tested the method using a “blind structure prediction” challenge. This self-assessment technique, responsible for the enormous progress in the world of protein structure modeling, allows scientists to test the fidelity of their computational models by predicting the three-dimensional structure of a known molecule and then comparing their proposed structure to the X-ray crystallography results.
The success in the test suggests that reliable structure prediction for complex three-dimensional folds is within reach, an enormous step forward on the path to reliable and efficient computational peptoid design.
“This will hopefully break open the field of peptoid structure prediction and design, an area we desperately need to guide our more well-developed synthetic efforts,” says Ron Zuckermann, co-author and director of the Biological Nanostructures Facility at Berkeley Lab’s Molecular Foundry.
Portions of this work were performed at the Molecular Foundry, Lawrence Berkeley National Laboratory, and supported by the DOE Office of Science. Additional funding came from the Defense Threat Reduction Agency, the NSF and the NIH.