Hacking life: how to program new functions for living bacteria and yeast
April 4, 2016

MIT biological engineers have devised a programming language that can be used to add new functions to E. coli bacteria (credit: Janet Iwasa)
MIT biological engineers have created a programming language for bacteria. It allows anyone to rapidly design complex, DNA-encoded circuits that add new functions to living cells — no genetic engineering knowledge required.
For example: design bacterial cells that can produce a cancer drug when they detect a tumor or create yeast cells that can halt their own fermentation process if too many toxic byproducts build up.
Repurposing a computer-chip programming language
The language is based on Verilog, a text-based language commonly used to program computer chips. To create a version of the language that would work for cells, the researchers designed computing elements such as logic gates and sensors that can be encoded in a bacterial cell’s DNA.
The sensors can detect different compounds, such as oxygen or glucose, and environmental conditions like light, temperature, and acidity. You can also add your own sensors, and you can compile a program for different organisms to get the right DNA sequence for each one.
You also specify the sensors, actuators, and a user constraints file (UCF), which defines the organism, gate technology, and valid operating conditions.
Cello (think of it as a compiler), a website, then uses this information to automatically design a DNA sequence encoding the desired circuit, using a set of algorithms that parse the Verilog text, create the circuit diagram, assign gates, balance constraints to build the DNA, and simulate performance, drawing on a library of Boolean logic gates.
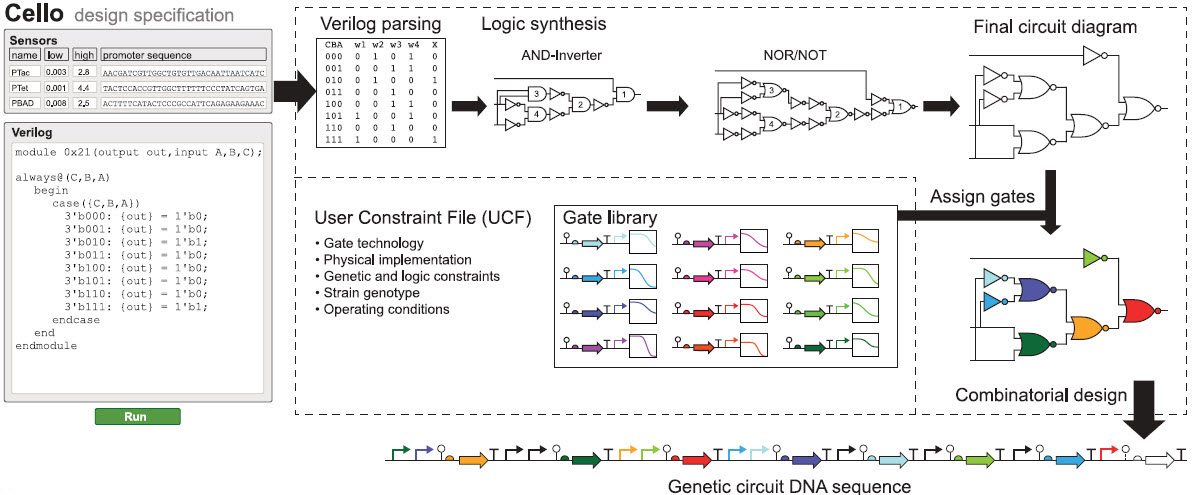
Overview of Cello. Cello users write Verilog code and select or upload sensors and a UCF. On the basis of the Verilog design, a truth table is constructed, from which a circuit diagram is synthesized. Regulators are assigned from a library to each gate (each color is a different repressor). Combinatorial design is then used to concatenate parts into a linear DNA sequence. (credit: Alec A. K. Nielsen et al./Science)
In the current Verilog version, the genetic parts are optimized for E. coli, but the researchers are working on expanding the language for other strains of bacteria, including Bacteroides, commonly found in the human gut; Pseudomonas,which often lives in plant roots; and the yeast Saccharomyces cerevisiae.
Christopher Voigt, an MIT professor of biological engineering, and colleagues at Boston University and the National Institute of Standards and Technology have used this language, which they describe in the April 1 issue of Science.
No experience needed
Over the past 15 years, biologists and engineers have designed many genetic parts, such as sensors, memory switches, and biological clocks, that can be combined to modify existing cell functions and add new ones. However, designing each circuit is a laborious process that requires great expertise and often a lot of trial and error. “You have to have this really intimate knowledge of how those pieces are going to work and how they’re going to come together,” Voigt says.
Users of the new programming language, however, need no special knowledge of genetic engineering.
“You could be completely naive as to how any of it works. That’s what’s really different about this,” Voigt says. “You could be a student in high school and go onto the Web-based server and type out the program you want, and it spits back the DNA sequence.”
Using this language, the MIT researchers programmed 60 circuits with different functions, and 45 of them worked correctly the first time they were tested. Many of the circuits were designed to measure one or more environmental conditions, such as oxygen level or glucose concentration, and respond accordingly. Another circuit was designed to rank three different inputs and then respond based on the priority of each one.
One of the new circuits is the largest biological circuit ever built, containing seven logic gates and about 12,000 base pairs of DNA.
Another advantage of this technique is its speed. Until now, “it would take years to build these types of circuits. Now you just hit the button and immediately get a DNA sequence to test,” Voigt says.
His team plans to work on several different applications using this approach: bacteria that can be swallowed to aid in digestion of lactose; bacteria that can live on plant roots and produce insecticide if they sense the plant is under attack; and yeast that can be engineered to shut off when they are producing too many toxic byproducts in a fermentation reactor.
Abstract of Genetic circuit design automation
Computation can be performed in living cells by DNA-encoded circuits that process sensory information and control biological functions. Their construction is time-intensive, requiring manual part assembly and balancing of regulator expression. We describe a design environment, Cello, in which a user writes Verilog code that is automatically transformed into a DNA sequence. Algorithms build a circuit diagram, assign and connect gates, and simulate performance. Reliable circuit design requires the insulation of gates from genetic context, so that they function identically when used in different circuits. We used Cello to design 60 circuits for Escherichia coli (880,000 base pairs of DNA), for which each DNA sequence was built as predicted by the software with no additional tuning. Of these, 45 circuits performed correctly in every output state (up to 10 regulators and 55 parts), and across all circuits 92% of the output states functioned as predicted. Design automation simplifies the incorporation of genetic circuits into biotechnology projects that require decision-making, control, sensing, or spatial organization.