On-line gamers succeed where scientists fail, opening door to new AIDS drug design
September 20, 2011
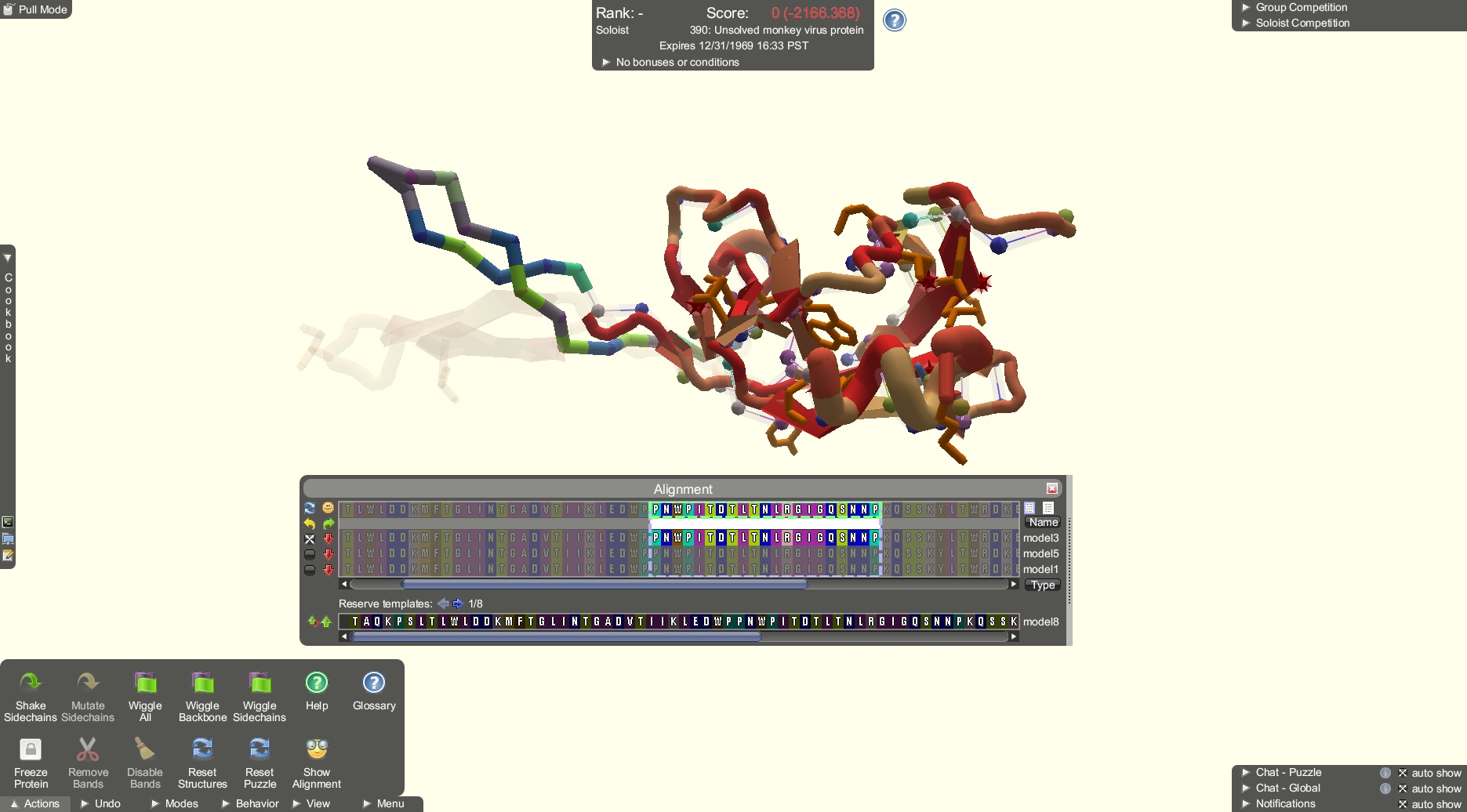
The "unsolved monkey virus protein" Foldit puzzle, highlighting the tool used by online gamers (credit: University of Washington)
Online gamers have solved the structure of a retrovirus enzyme whose configuration had stymied scientists, biochemist Firas Khatib of the University of Washington (UW) reports.
The players were adept at a computer game, Foldit, that allows players to collaborate and compete in predicting the structure of protein molecules.
After scientists repeatedly failed to piece together the structure of a protein-cutting enzyme from an AIDS-like virus, they called in the Foldit players and challenged them to produce an accurate model of the enzyme.
Remarkably, the gamers generated models good enough for the researchers to refine and determine the enzyme’s structure within a few days. Equally amazing, surfaces on the molecule stood out as likely targets for drugs to deactivate the enzyme.
This class of enzymes, called retroviral proteases, has a critical role in how the AIDS virus matures and proliferates. Intensive research is underway to try to find anti-AIDS drugs that can block these enzymes, but efforts were hampered by not knowing exactly what the retroviral protease molecule looked like.
“These features provide exciting opportunities for the design of retroviral drugs, including AIDS drugs,” write Khatib and co-authors (including the gamers) of a paper appearing yesterday in the journal Nature Structural & Molecular Biology.
“Online gamers have solved a longstanding scientific problem, perhaps leading the way to new anti-viral drugs,” said Carter Kimsey, program director in the National Science Foundation’s (NSF) Division of Biological Infrastructure, which funded the research.
Fold-it was created by computer scientists and biochemists at the UW Center for Game Science, and by paper co-author biochemist David Baker of UW, to engage the general public in scientific discovery.
The solution of the virus enzyme structure, the scientists said, indicates the power of online computer games to channel human intuition and of three-dimensional pattern-matching skills to solve challenging scientific problems. The online protein folding game captivates thousands of avid players worldwide.
Direct manipulation tools, as well as assistance from a computer program called Rosetta, encourage participants to configure graphics into a workable protein model. Teams send in their answers, and UW researchers constantly improve the design of the game and its puzzles by analyzing the players’ problem-solving strategies.
Figuring out the shape and misshape of proteins contributes to research on causes of and cures for cancer, Alzheimer’s, immune deficiencies, and a host of other disorders, as well as to environmental work on biofuels.
Games like Foldit are evolving. To piece together the retrovirus enzyme structure, Cooper said, Foldit used a new alignment tool for the first time to copy parts of known molecules and test their fit in an incomplete model.
“The ingenuity of game players,” the paper concludes, “is a formidable force that, if properly directed, can be used to solve a wide range of scientific problems.”
Ref.: Firas Khatib, et al., Crystal structure of a monomeric retroviral protease solved by protein folding game players, Nature Structural and Molecular Biology, 2011; [DOI:10.1038/nsmb.2119]