Novel DNA origami structures
July 24, 2015

The versatility of the DNA origami 3D wireframe design technique created by Arizona State University Biodesign Institute researcher Hao Yan is demonstrated with the construction of this snub cube model, an Archimedean solid with 60 edges, 24 vertices and 38 faces including 6 squares and 32 equilateral triangles. (credit: TED-43/Wikimedia Commons)
Hao Yan, a researcher at Arizona State University’s Biodesign Institute, has extended DNA origami — which uses combinations of DNA base pairs to create new 2-D and 3-D nanoforms — into imaginative new forms that may one day lead to microelectronics and biomedical innovations.
“Earlier design methods [for DNA origami] used strategies including parallel arrangement of DNA helices to approximate arbitrary shapes, but precise fine-tuning of DNA wireframe architectures that connect vertices in 3D space has required a new approach,” says Yan, the Milton D. Glick Distinguished Chair of Chemistry and Biochemistry at ASU and directs Biodesign’s Center for Molecular Design and Biomimetics.
The new study describes wireframe structures of high complexity and programmability that are fabricated by precise control of branching and curvature, using novel organizational principles for the designs. (Wireframes are skeletal three-dimensional models represented purely through lines and vertices.)
The resulting nanoforms include symmetrical lattice arrays, quasicrystalline structures, curvilinear arrays, and a simple wire art sketch in the 100-nm scale, as well as 3-D objects including a snub cube with 60 edges and 24 vertices and a reconfigurable Archimedean solid that can be controlled to make the unfolding and refolding transitions between 3D and 2D.
The research appears in the advanced online edition of the journal Nature Nanotechnology.
DNA origami: how it works
In previous investigations, the Yan group created subtle architectural forms at an astonishingly minute scale, some measuring only tens of nanometers across — roughly the diameter of a virus particle. These nano-objects include spheres, spirals, flasks, Möbius forms, and even an autonomous spider-like robot capable of following a prepared DNA track.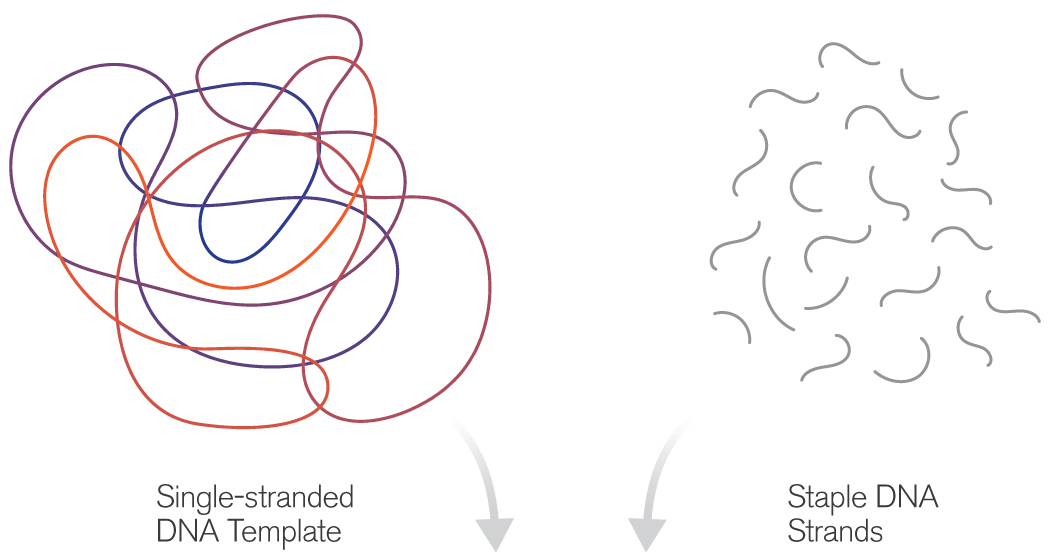
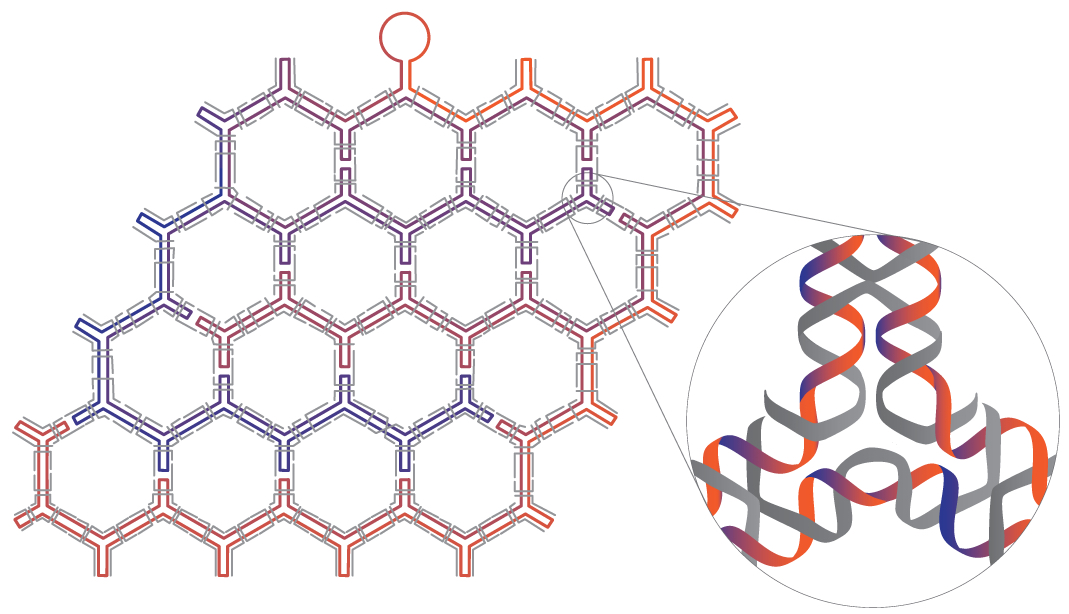
(credit: Fei Zhang et al./Nature Nanotechnology)
The technique of DNA origami capitalizes on the simple base-pairing properties of DNA, a molecule built from the four nucleotides Adenine (A), Thymine (T), Cytosine (C) and Guanine (G). The rules of the game are simple: A’s always pair with T’s and C’s with G’s. Using this abbreviated vocabulary, the myriad body plans of all living organisms are constructed; though duplicating even Nature’s simpler designs has required great ingenuity.
The basic idea of DNA origami is to use a length of single-stranded DNA as a scaffold for the desired shape. Base-pairing of complementary nucleotides causes the form to fold and self-assemble. The process is guided by the addition of shorter “staple strands,” which act to help fold the scaffold and to hold the resulting structure together. Various imaging technologies are used to observe the tiny structures, including fluorescence-, electron- and atomic force microscopy.
DNA origami originally produced nanoarchitectures of purely aesthetic interest, but refinements of the technique have opened the door to a new range of exciting applications including molecular cages for the encapsulation of molecules, enzyme immobilization and catalysis, chemical and biological sensing tools, drug delivery mechanisms, and molecular computing devices.
The technique described in the new study takes this approach a step further, allowing researchers to overcome local symmetry restrictions, creating wireframe architectures with higher order arbitrariness and complexity. Here, each line segment and vertex is individually designed and controlled. The number of arms emanating from each vertex may be varied from 2 to 10 and the precise angles between adjacent arms can be modified.
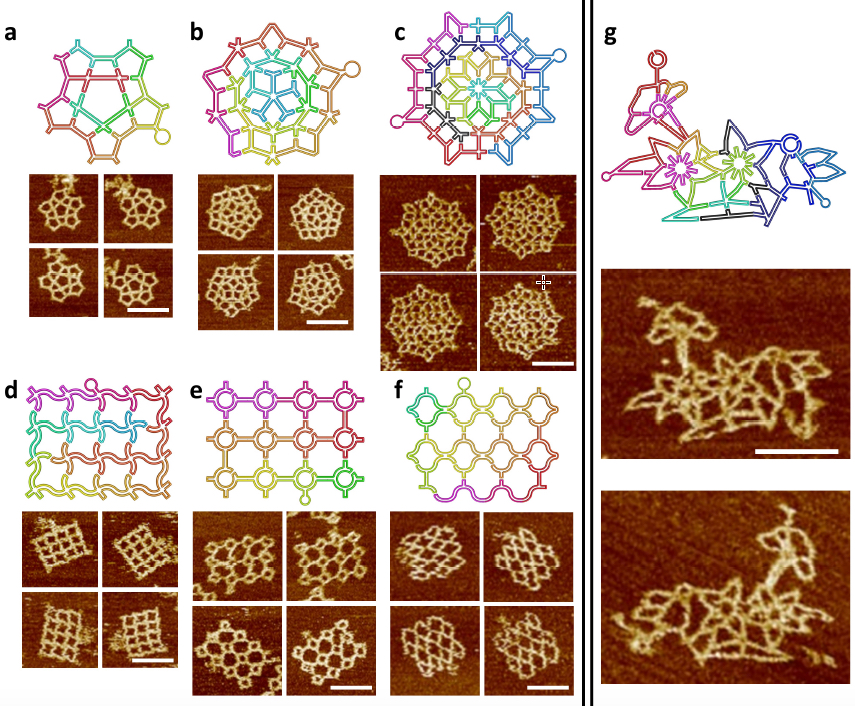
The scaffold folding path and representative AFM images for intricate 2D patterns. (a) A star-shape pattern without translational symmetry. (a) A Penrose tiling. (c) An 8-fold quasicrystalline pattern. (d-f) Three curved structures. (d) A waving grid. (e) A sphere array. (f) A fishnet. (g) A flower-and-bird pattern. All scale bars are 100 nm. Two scaffolds (one colorful and one black-blue) are used in c and g. (credit: Fei Zhang et al./Nature Nanotechnology)
In the current study, the method was first applied to symmetrical, regularly repeating polygonal designs, including hexagonal, square and triangular tiling geometries. Such common designs are known as tessellation patterns. A clever strategy involving a series of bridges and loops was used to properly route the scaffold strand, allowing it to pass through the entire structure, touching all lines of the wireframe once and only once. Staple strands were then applied to complete the designs.
In subsequent stages, the researchers created more complex wireframe structures, without the local translational symmetry found in the tessellation patterns. Three such patterns were made, including a star shape, a 5-fold Penrose tile and an 8-fold quasicrystalline pattern. (Quasicrystals are structures that are highly ordered but non-periodic. Such patterns can continuously fill available space, but are not translationally symmetric.)
Loop structures inserted into staple strands and unpaired nucleotides at the vertex points of the scaffold strands were also used, allowing researchers to perform precision modifications to the angles of junction arms.
The new design rules were next tested with the assembly of increasingly complex nanostructures, involving vertices ranging from 2 to 10 arms, with many different angles and curvatures involved, including a complex pattern of birds and flowers. The accuracy of the design was subsequently confirmed by AFM imaging, proving that the method could successfully yield highly sophisticated wireframe DNA nanostructures.
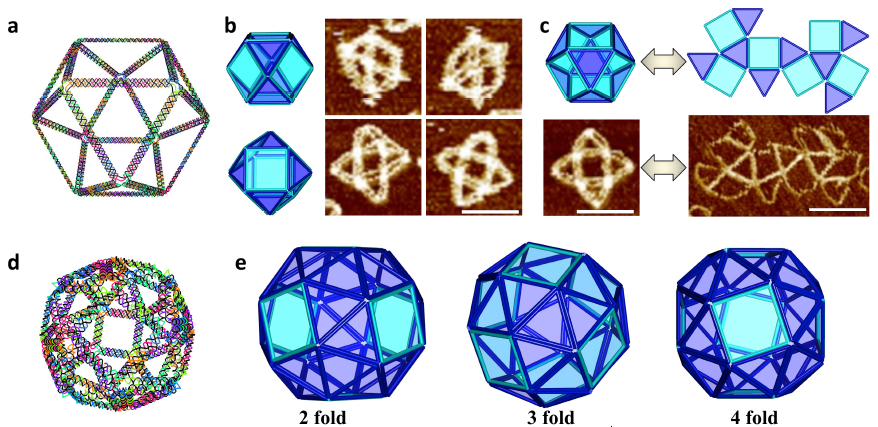
3D wire-frame Archimedean solid structures. (a) A 3D model of an Archimedean solid cuboctahedron with 12 vertices and 24 edges. Each vertex is a 4 Å~ 4 junction, and each edge is a 14-turn long double DNA duplex. (b) Left: Models showing possible conformations of the structure when deposited on mica surface; Right: the corresponding AFM images. (c) The reconfiguration between 3D and 2D can be realized by strand displacement by adding fuel and set strands. Top: reconfiguration schematics, Bottom: AFM images showing the transition. All scale bars in AFM images are 100 nm. (d) A 3D model of snub cube with 24 vertices and 60 edges. Each vertex is 5 Å~ 4 junction, and each edge is a 5-turn double DNA duplex. (e) Three views of the DNA snub cube from the design model (top of page). (credit: Fei Zhang et al./Nature Nanotechnology)
The method was then adapted to produce a number of 3D structures as well, including a cuboctahedron, and another Archimedian solid known as a snub cube — a structure with 60 edges, 24 vertices and 38 faces, including 6 squares and 32 equilateral triangles.
The authors stress that the new design innovations described can be used to compose and construct any imaginable wireframe nanostructure— a significant advancement for the burgeoning field.
On the horizon, nanoscale structures may one day be marshaled to hunt cancer cells in the body or act as robot assembly lines for the design of new drugs.
Abstract of Complex wireframe DNA origami nanostructures with multi-arm junction vertices
Structural DNA nanotechnology and the DNA origami technique, in particular, have provided a range of spatially addressable two- and three-dimensional nanostructures. These structures are, however, typically formed of tightly packed parallel helices. The development of wireframe structures should allow the creation of novel designs with unique functionalities, but engineering complex wireframe architectures with arbitrarily designed connections between selected vertices in three-dimensional space remains a challenge. Here, we report a design strategy for fabricating finite-size wireframe DNA nanostructures with high complexity and programmability. In our approach, the vertices are represented by n × 4 multi-arm junctions (n = 2–10) with controlled angles, and the lines are represented by antiparallel DNA crossover tiles of variable lengths. Scaffold strands are used to integrate the vertices and lines into fully assembled structures displaying intricate architectures. To demonstrate the versatility of the technique, a series of two-dimensional designs including quasi-crystalline patterns and curvilinear arrays or variable curvatures, and three-dimensional designs including a complex snub cube and a reconfigurable Archimedean solid were constructed.