Scientists discover precise DNA sequence code critical for turning genes on
January 27, 2017
DNA sequence signal for the activation of human genes. Each tiny human cell contains about six feet of DNA, a double-helical molecular chain containing four types of several billion chemical nucleotides — adenine (A), cytosine (C), guanine (G) and thymine (T) — arranged in a specific sequence, or code, that when transcribed guide the cell into producing specific proteins. (credit: University of California — San Diego)
Molecular biologists at the University of California, San Diego (UC San Diego) have discovered a short sequence of DNA that is essential for turning on (expressing proteins) more than half of all human genes — an achievement that should provide scientists with a better understanding of how human genes are regulated.
Knowing what turns on genes is important. Each human cell contains about six feet of DNA, a double-helical molecular chain containing several billion chemical nucleotides — adenine (A), cytosine (C), guanine (G) and thymine (T) — arranged in a specific sequence, or code. Active genes undergo a process called transcription, in which the nucleotide sequence in DNA is read and converted into a sister language called RNA for processing. The processed RNA sequence then guides the cell to produce specific proteins that are essential for normal cellular functions.

A depiction of the double helical structure of DNA. Its four nucleotide coding units (A, T, C, G) are color-coded in pink, orange, purple and yellow. (credit: NHGRI)
“In these six feet of DNA, there are tens of thousands of genes, which are segments of DNA that direct specific functions, such as the production of a hormone or an enzyme,” explains James T. Kadonaga, PhD, a molecular biology professor at UC San Diego who headed the team of researchers. “It is essential for the cell to control the activity of each of its tens of thousands of genes, because the improper control of gene activity can lead to adverse outcomes such as cell death or the formation of a cancer cell.”
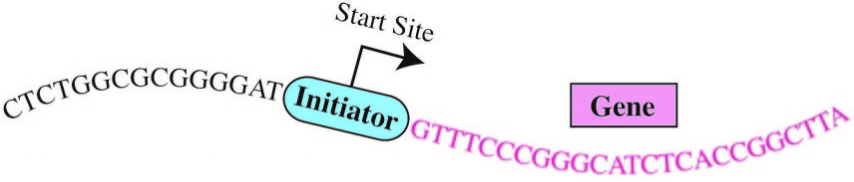
The “human Initiator”
Enter the “Initiator.” The initiation of gene expression often occurs at a critical DNA sequence code called the “human Initiator.” This small piece of DNA helps gene expression machinery locate exactly where to begin transcribing. Although the concept of the Initiator has been known since the 1980s, the precise DNA sequence comprising the Initiator had eluded scientists.*
“There are many sequence signals that control gene activity in human cells and the Initiator is the most commonly occurring sequence at the start sites of genes,” Kadonaga said. Kadonaga and his team employed emerging genomic techniques and devised novel computational strategies to unlock the exact DNA sequence code for the human Initiator.
They also discovered that this sequence is located precisely at the start site of more than half of all human genes, underlining the importance of the human Initiator in the human genome. “The solution of the human Initiator code will enable us to explore new frontiers in gene regulation,” said Kadonaga. “In the future, it will be possible to use the code to identify other regulatory signals and in this way gain a more complete understanding of how human genes are turned on and off.”
“The authors verified the Initiator sequence in multiple cell lines, which is an impressive finding,” a scientist not involved in the studies told KurzweilAI. “However, none of these cell lines reflect normal human biology — they are essentially cancer cells proliferating in a dish. I would have liked to see this Initiator sequence verified in normal human cells from healthy patients.”
The research, now online and to be detailed in the February 10 print issue of the journal Genes & Development, was supported by grants from the National Institutes of Health.
* First observed by Pierre Chambon and his colleagues in Strasbourg, France in 1980, the human Initiator and its role in gene activation were articulated in 1989 by two MIT biologists, Stephen Smale and David Baltimore at MIT, who later revealed the approximate sequence code of the Initiator. Since then, however, other scientists had proposed a number of different sequences for the human Initiator, but none of them were found to be consistently associated with the start sites of human genes. As a result, the true Initiator sequence code remained a mystery — until now.
Abstract of The human initiator is a distinct and abundant element that is precisely positioned in focused core promoters
DNA sequence signals in the core promoter, such as the initiator (Inr), direct transcription initiation by RNA polymerase II. Here we show that the human Inr has the consensus of BBCA+1BW at focused promoters in which transcription initiates at a single site or a narrow cluster of sites. The analysis of 7678 focused transcription start sites revealed 40% with a perfect match to the Inr and 16% with a single mismatch outside of the CA+1 core. TATA-like sequences are underrepresented in Inr promoters. This consensus is a key component of the DNA sequence rules that specify transcription initiation in humans.