‘Tree of life’ for 2.3 million species released
September 21, 2015
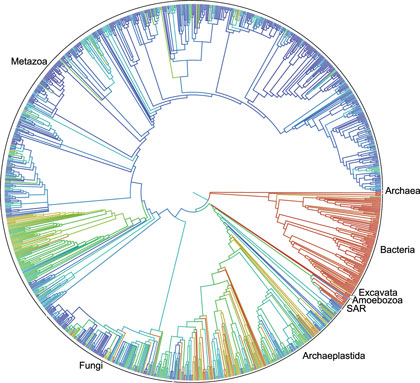
This circular family tree of Earth’s lifeforms is considered a first draft of the 3.5-billion-year history of how life evolved and diverged (credit: Duke University)
A first draft of the “tree of life” for the roughly 2.3 million named species of animals, plants, fungi and microbes — from platypuses to puffballs — has been released.
A collaborative effort among eleven institutions, the tree depicts the relationships among living things as they diverged from one another over time, tracing back to the beginning of life on Earth more than 3.5 billion years ago.
Tens of thousands of smaller trees have been published over the years for select branches of the tree of life — some containing upwards of 100,000 species — but this is the first time those results have been combined into a single tree that encompasses all of life.
“This is the first real attempt to connect the dots and put it all together,” said principal investigator Karen Cranston of Duke University. “Think of it as Version 1.0.” The current version of the tree — along with the underlying data and source code — is available to browse, edit, and download free at https://tree.opentreeoflife.org — a sort of “Wikipedia” for the evolutionary trees.
It is also described in an open-access article appearing Sept. 18 in the Proceedings of the National Academy of Sciences.
Uses of evolutionary trees
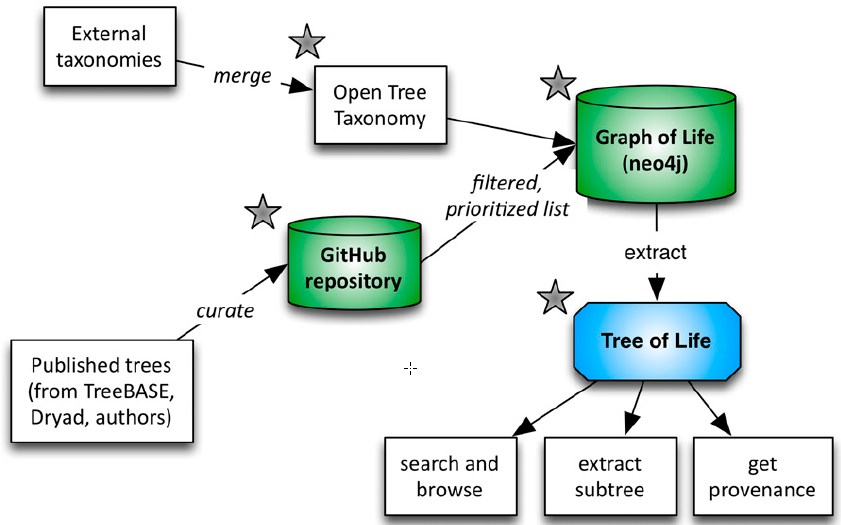
Open Tree of Life workflow (credit: Cody E. Hinchliff et al./PNAS)
Understanding how the millions of species on Earth are related to one another helps scientists discover new drugs, increase crop and livestock yields, and trace the origins and spread of infectious diseases such as HIV, Ebola and influenza, the scientists say.
The researchers pieced it together by compiling thousands of smaller chunks that had already been published online and merging them together into a gigantic “supertree” that encompasses all named species. The initial draft is based on nearly 500 smaller trees from previously published studies.
To map trees from different sources to the branches and twigs of a single supertree, one of the biggest challenges was simply accounting for the name changes, alternate names, common misspellings and abbreviations for each species. The eastern red bat, for example, is often listed under two scientific names, Lasiurus borealis and Nycteris borealis. Spiny anteaters once shared their scientific name with a group of moray eels.
“Although a massive undertaking in its own right, this draft tree of life represents only a first step,” the researchers wrote. For one, only a tiny fraction of published trees are digitally available.
A survey of more than 7,500 phylogenetic studies published between 2000 and 2012 in more than 100 journals found that only one out of six studies had deposited their data in a digital, downloadable format that the researchers could use.
The vast majority of evolutionary trees are published as PDFs and other image files that are impossible to enter into a database or merge with other trees.
As a result, the relationships depicted in some parts of the tree, such as the branches representing the pea and sunflower families, don’t always agree with expert opinion.
Other parts of the tree, particularly insects and microbes, remain elusive. That’s because even the most popular online archive of raw genetic sequences — from which many evolutionary trees are built — contains DNA data for less than five percent of the tens of millions species estimated to exist on Earth.
“As important as showing what we do know about relationships, this first tree of life is also important in revealing what we don’t know,” said co-author Douglas Soltis of the University of Florida.
To help fill in the gaps, the team is also developing software that will enable researchers to log on and update and revise the tree as new data come in for the millions of species still being named or discovered.
“It’s by no means finished,” Cranston said. “It’s critically important to share data for already-published and newly-published work if we want to improve the tree.”
“Twenty five years ago people said this goal of huge trees was impossible,” Soltis said. “The Open Tree of Life is an important starting point that other investigators can now refine and improve for decades to come.”
Abstract of Synthesis of phylogeny and taxonomy into a comprehensive tree of life
Reconstructing the phylogenetic relationships that unite all lineages (the tree of life) is a grand challenge. The paucity of homologous character data across disparately related lineages currently renders direct phylogenetic inference untenable. To reconstruct a comprehensive tree of life, we therefore synthesized published phylogenies, together with taxonomic classifications for taxa never incorporated into a phylogeny. We present a draft tree containing 2.3 million tips—the Open Tree of Life. Realization of this tree required the assembly of two additional community resources: (i) a comprehensive global reference taxonomy and (ii) a database of published phylogenetic trees mapped to this taxonomy. Our open source framework facilitates community comment and contribution, enabling the tree to be continuously updated when new phylogenetic and taxonomic data become digitally available. Although data coverage and phylogenetic conflict across the Open Tree of Life illuminate gaps in both the underlying data available for phylogenetic reconstruction and the publication of trees as digital objects, the tree provides a compelling starting point for community contribution. This comprehensive tree will fuel fundamental research on the nature of biological diversity, ultimately providing up-to-date phylogenies for downstream applications in comparative biology, ecology, conservation biology, climate change, agriculture, and genomics.