Single-cell RNA sequencing yields genetic makeup of human and mouse embryos in unprecedented detail
August 2, 2013
UCLA scientists, in collaboration with teams in China, have used the powerful technology of single-cell RNA sequencing to track the genetic development of a human and a mouse embryo at an unprecedented level of accuracy.
The technique could lead to earlier and more accurate diagnoses of genetic diseases — even when the embryo consists of only eight cells.
The study was led by Guoping Fan, professor of human genetics and molecular biology and member of both the Jonsson Comprehensive Cancer Center and the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research.
Single-cell RNA sequencing allows researchers to determine the precise nature of the total gene transcripts, or all of the genes that are actively expressed in a particular cell.
“The advantages of this technique are twofold,” Fan said. “It is a much more comprehensive analysis than was achievable before and the technique requires a very minimal amount of sample material — just one cell.”
Major development in genetic diagnoses
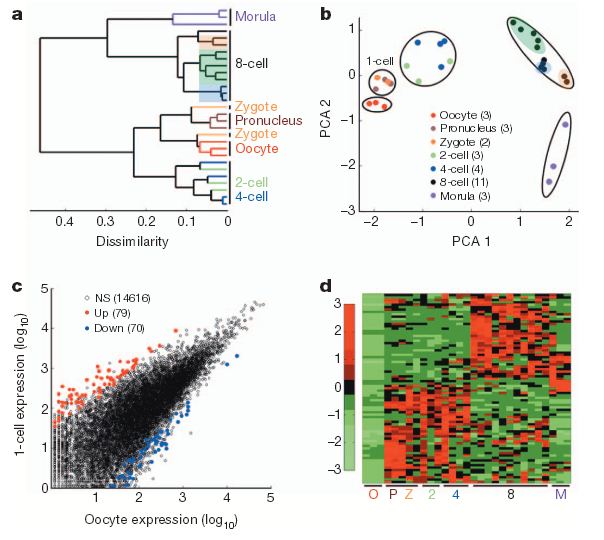
High-resolution single-cell transcriptome analysis of human pre-implantation embryos. a, Unsupervised hierarchical clustering. b, Principal component analysis of single blastomere expression patterns for seven stages of oocytes and pre-implantation embryos. Sister cells from the same 8-cell embryo are highlighted together. The number of samples for each stage is indicated in the PCA legend in parenthesis. c, Scatterplot showing the number of activated (red) and reduced (blue) genesin 1-cell embryos (n55) compared to oocytes (n53). d, Heatmap showing relative expression patterns of 1-cell activated genes (n579) across all pre-implantation stages. NS, not significant. (Credit: Zhigang Xue et al./Nature)
Besides its implications for genetic diagnoses — such as improving scientists’ ability to identify genetic mutations like BRCA1 and BRCA2, which predispose women to breast cancer and ovarian cancer, or genetic diseases that derive from protein dysfunction, such as sickle cell disease — the technology may also have important uses in reproductive medicine.
The technique marks a major development in genetic diagnoses, which previously could not be conducted this early in embryonic development and required much larger amounts of biological material.
“Previous to this paper we did not know this much about early human development,” said Kevin Huang, the study’s co-first author and a postdoctoral scholar in Fan’s laboratory. “Now we can define what ‘normal’ looks like, so in the future we will have a baseline from which to compare possible genetic problems. This is our first comprehensive glance at what is normal.”
With single-cell RNA sequencing, much more gene transcription was detected than before. “The question we asked is, ‘How does the gene network drive early development from one cell to two cells, two cells to four cells, and so on?'” Fan said. “Using the genome data analysis methods developed by co-author Steve Horvath at UCLA, we have uncovered crucial gene networks and we can now predict possible future genetic disorders at the eight-cell stage.”
The research was supported by the Chinese Ministry of Science and Technology, the International Science and Technology Cooperation Program of China, and the National Natural Science Foundation of China.