Why you’re smarter than a chicken
August 21, 2015

Sorry, wrong protein — you’re dinner (credit: Johnathan Nightingale via Flickr)
A single molecular event in a protein called PTBP1 in our cells could hold the key to how we evolved to become the smartest animal on the planet, University of Toronto researchers have discovered.
The conundrum: Humans and frogs, for example, have been evolving separately for 350 million years and use a remarkably similar repertoire of genes to build organs in the body. So what accounts for the vast range of organ size and complexity?
Benjamin Blencowe, a professor in the University of Toronto’s Donnelly Centre and Banbury Chair in Medical Research, and his team believe they now have the key: alternative splicing (AS).
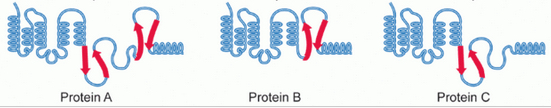
With alternative splicing, the same gene can generate three different types of protein molecules, as in this example (credit: Wikipedia)
Here’s how alternative splicing works: specific sections of a gene called exons may be included or excluded from the final messenger RNA (mRNA) that expresses the gene (creates proteins). And that changes the arrangement of amino acid sequences.
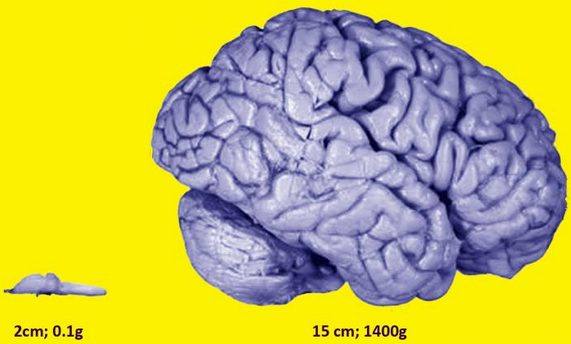
This image shows a frog and human brain, brought to scale. Although the brain-building genes are similar in both, alternative splicing ensures greater protein diversity in human cells, which fuels organ complexity. (credit: Jovana Drinjakovic)
There are two forms of PTBP1: one that is common in all vertebrates, and another in mammals. The researchers showed that in mammalian cells, the presence of the mammalian version of PTBP1 unleashes a cascade of alternative splicing events that lead to a cell becoming a neuron instead of a skin cell, for example.
To prove that, they engineered chicken cells to make mammalian-like PTBP1, and this triggered alternative splicing events that are found in mammals, creating a smart chicken (no relation to the eponymous brand). Also, in turns out that alternative splicing prevalence increases with vertebrate complexity.
The end result: all those small accidental changes across specific genes have fueled the evolution of mammalian brains.
The study is published in the August 20 issue of Science.
Abstract of An alternative splicing event amplifies evolutionary differences between vertebrates
Alternative splicing (AS) generates extensive transcriptomic and proteomic complexity. However, the functions of species- and lineage-specific splice variants are largely unknown. Here we show that mammalian-specific skipping of polypyrimidine tract–binding protein 1 (PTBP1) exon 9 alters the splicing regulatory activities of PTBP1 and affects the inclusion levels of numerous exons. During neurogenesis, skipping of exon 9 reduces PTBP1 repressive activity so as to facilitate activation of a brain-specific AS program. Engineered skipping of the orthologous exon in chicken cells induces a large number of mammalian-like AS changes in PTBP1 target exons. These results thus reveal that a single exon-skipping event in an RNA binding regulator directs numerous AS changes between species. Our results further suggest that these changes contributed to evolutionary differences in the formation of vertebrate nervous systems.