A self-assembling protein nanocage
June 10, 2014
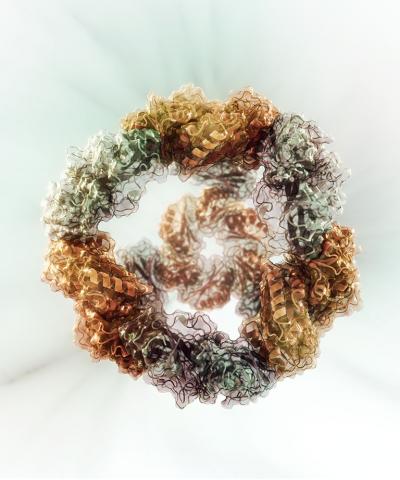
A computational model of a successfully designed two-component protein nanocage with tetrahedral symmetry that could one day be used to deliver cancer-drug molecules (credit: Dr. Vikram Mulligan)
University of Washington (UW) scientists have developed a new computational method for building new customized proteins that self-assemble (like biological systems) to revolutionize things like targeted delivery of drugs, vaccine development, and even electronic devices.
The work is based in the Rosetta macromolecular modeling package, which was developed by David Baker’s laboratory at the UW Institute for Protein Design, in collaboration with colleagues at UCLA and Janelia Farm.
The program was originally created to predict natural protein structures from amino acid sequences. Researchers are increasingly using Rosetta to design new protein structures and sequences aimed at solving real-world problems.
Reporting in the June 5 issue of Nature, the researchers describe the development and application of new Rosetta software enabling the design of novel protein nanomaterials composed of multiple copies of distinct protein subunits, which arrange themselves into higher order, symmetrical architectures.
A cage for delivering anti-cancer drugs
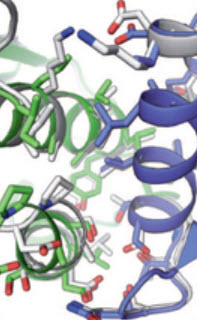
Visualization of a self-assembling protein nanostructure in process (credit: King, N., Bale, J.B., Sheffler, W., et al. Nature)
With the new software, the scientists were able to create five novel, 24-subunit cage-like protein nanomaterials. Importantly, the actual structures, the researchers observed, were in very close agreement with their computer modeling.
Their method depends on encoding pairs of protein amino acid sequences with the information needed to direct molecular assembly through protein-protein interfaces.
The interfaces not only provide the energetic forces that drive the assembly process, they also precisely orient the pairs of protein building blocks with the geometry required to yield the desired cage-like symmetric architectures.
Creating this cage-shaped protein, the scientists said, may be a first step towards building nanoscale containers.
According to UW translational investigator Neil King, who leads the new research, he looks forward to a time when cancer-drug molecules will be packaged inside of designed nanocages and delivered directly to tumor cells, sparing healthy cells.
(See previous KurzweilAI coverage of anti-cancer drug-delivery research here.)
The scientists note that combining just two types of symmetry elements, as in this study, can in theory give rise to a range of symmetrical shapes, such as cubic point groups, helices, layers, and crystals.

Detail of computational model of a successfully designed two-component protein nanocage with tetrahedral symmetry (rendering by Vikram Mulligan)
King explained that the immune system responds to repetitive, symmetric patterns, such as those on the surface of a virus or disease bacteria. Building nano-decoys may be a way train the immune system to attack certain types of pathogens.
From vaccines to clean energy
“This concept may become the foundation for vaccines based on engineered nanomaterials,” King said. Further down the road, he and Bale anticipate that these design methods can also be used for developing new clean-energy technologies.
The scientists added in their report, “The precise control over interface geometry offered by our method enables the design of two-component protein nanomaterials with diverse nanoscale features, such as surfaces, pores, and internal volumes, with high accuracy.”
They said the combinations possible with two-component materials greatly expand the number and variety of potential nanomaterials that could be designed. It may even may be possible to produce nanomaterials in a variety of sizes, shapes and arrangements, and also move on to construct increasingly more complex materials from more than two components, and even be programmable, they said.
This project was supported with funding from the Howard Hughes Medical Institute, the National Science Foundation, the International Vaccine Initiative, the U.S. Air Force Office of Scientific Research, and the U.S. Department of Energy.
Abstract of Nature paper
The self-assembly of proteins into highly ordered nanoscale architectures is a hallmark of biological systems. The sophisticated functions of these molecular machines have inspired the development of methods to engineer self-assembling protein nanostructures; however, the design of multi-component protein nanomaterials with high accuracy remains an outstanding challenge. Here we report a computational method for designing protein nanomaterials in which multiple copies of two distinct subunits co-assemble into a specific architecture. We use the method to design five 24-subunit cage-like protein nanomaterials in two distinct symmetric architectures and experimentally demonstrate that their structures are in close agreement with the computational design models. The accuracy of the method and the number and variety of two-component materials that it makes accessible suggest a route to the construction of functional protein nanomaterials tailored to specific applications.