Credit card-sized device could analyze biopsy, help diagnose pancreatic cancer in minutes
March 6, 2014
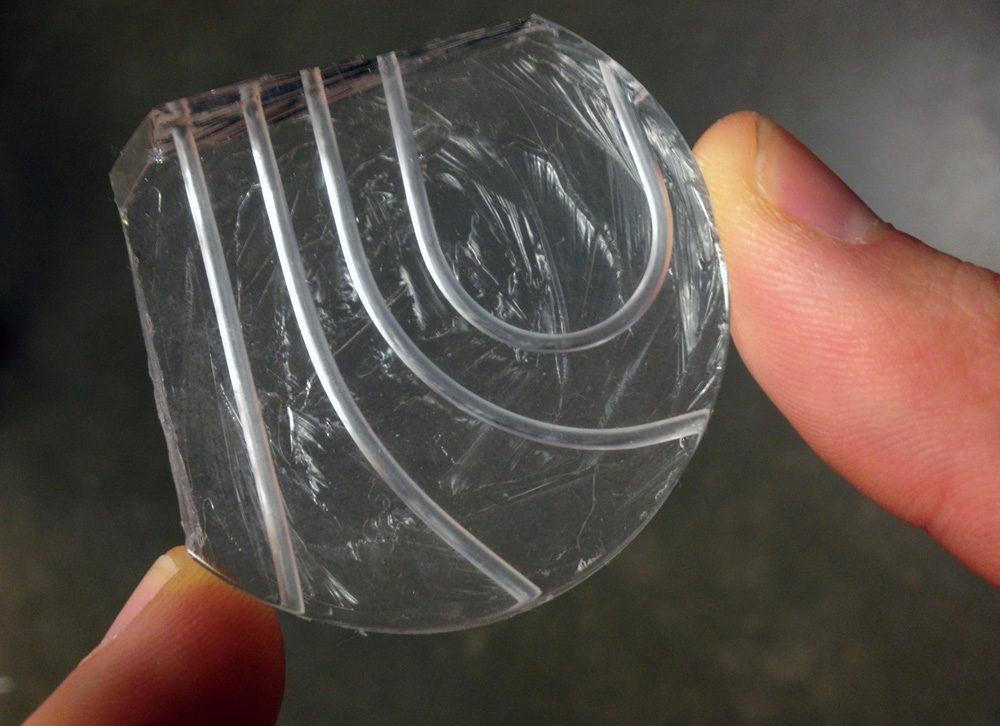
This prototype of a microfluidic device has both curved and straight channels for transporting tissue biopsies. The silicon material is lightweight, flexible and transparent (credit: University of Washington)
University of Washington scientists and engineers are developing a low-cost device that could help pathologists diagnose pancreatic cancer* earlier and faster.
The prototype can perform the basic steps for processing a biopsy, relying on fluid transport instead of human hands to process the tissue.
“This new process is expected to help the pathologist make a more rapid diagnosis and be able to determine more accurately how invasive the cancer has become, leading to improved prognosis,” said Eric Seibel, a UW research professor of mechanical engineering and director of the department’s Human Photonics Laboratory.
Currently, a pathologist takes a biopsy tissue sample, then sends it to the lab where it’s cut into thin slices, stained and put on slides, then analyzed optically in 2D for abnormalities.
The UW’s technology would process and analyze whole tissue biopsies for 3D imaging, which offers a more complete picture of the cellular makeup of a tumor, said Ronnie Das, a UW postdoctoral researcher in bioengineering, the lead author on a related paper.
“As soon as you cut a piece of tissue, you lose information about it. If you can keep the original tissue biopsy intact, you can see the whole story of abnormal cell growth. You can also see connections, cell morphology and structure as it looks in the body,” Das said.
Microfluidics for tissue
The research team is building a thick, credit card-sized, flexible device out of silicon that allows a piece of tissue to pass through tiny channels and undergo a series of steps that replicate what happens on a much larger scale in a pathology lab.
The device uses microfluidics, which allows tissue to move and stop with ease through small channels without needing to apply a lot of external force. It also keeps clinicians from having to handle the tissue; instead, a tissue biopsy taken with a syringe needle could be deposited directly into the device to begin processing.
The researchers say this is the first time material larger than a single-celled organism has successfully moved in a microfluidic device. This could have implications across the sciences in automating analyses that usually are done by humans, according to the researchers.
The device was designed to be simple to manufacture and use. They first built a mold using a petri dish and Teflon tubes, then poured a viscous, silicon material into the mold. The result is a small, transparent instrument with seamless channels that are both curved and straight.
The UW researchers say the technology could be used overseas as an over-the-counter kit that would process biopsies, then send that information to pathologists who could look for signs of cancer from remote locations. Additionally, it could potentially reduce the time it takes to diagnose cancer to a matter of minutes, Das said.
Next steps
The researchers have used the instrument to process a tissue biopsy one step at a time, following the same steps as a pathology lab would. Next, they hope to combine all of the steps into a more robust device — including 3D imaging — then build and optimize it for use in a lab. Future iterations of the device could include layers of channels that would allow more analyses on a piece of tissue without adding more bulk to the device.
“There are no other microfluidics devices designed specifically for needle biopsy preparation and analysis by a pathologist,” Seibel told KurzweilAI. “Every microfluidic device that is known to the researchers is designed for cells or sub-cellular components like DNA & proteins.
“Other imaging methodologies can image needle biopsy specimens rapidly in 3D, but no other system is a direct extension of absorption imaging based on staining the tissue while also allowing fluorescence molecular-specific imaging.
“Currently this is an exploratory project in academia, funded by only a small grant from the US National Science Foundation. However, there is great commercial potential in more rapidly moving pathology from preparing specimens by hand, slicing up tissue for creating thin flat images, to automating the specimen preparation process and imaging the entire biopsy specimen while preserving the tissue architecture.
“Our first application is for pancreatic tissue biopsy, where tissue architecture is thought to be very important in better understanding the cancer state and to develop a management plan for the patient.”
The team presented its initial results at the February 2014 SPIE Photonics West conference and recently filed a patent for this first-generation device and future technology advancements.
The research is funded by the National Science Foundation Bioengineering division and the U.S. Department of Education Graduate Assistance in Areas of National Need program.
* Pancreatic cancer is a particularly devastating disease. At least 94 percent of patients will die within five years, and in 2013 it was ranked as one of the top 10 deadliest cancers. Routine screenings for breast, colon and lung cancers have improved treatment and outcomes for patients with these diseases, largely because the cancer can be detected early. But because little is known about how pancreatic cancer behaves, patients often receive a diagnosis when it’s already too late.
Abstract of Proceedings SPIE paper
The pancreas is a deeply seated organ requiring endoscopically, or radiologically guided biopsies for tissue diagnosis. Current approaches include either fine needle aspiration biopsy (FNA) for cytologic evaluation, or core needle biopsies (CBs), which comprise of tissue cores (L = 1-2 cm, D = 0.4-2.0 mm) for examination by brightfield microscopy. Between procurement and visualization, biospecimens must be processed, sectioned and mounted on glass slides for 2D visualization. Optical information about the native tissue state can be lost with each procedural step and a pathologist cannot appreciate 3D organization from 2D observations of tissue sections 1-8 μm in thickness. Therefore, how might histological disease assessment improve if entire, intact CBs could be imaged in both brightfield and 3D? CBs are mechanically delicate; therefore, a simple device was made to cut intact, simulated CBs (L = 1-2 cm, D = 0.2-0.8 mm) from porcine pancreas. After CBs were laid flat in a chamber, z-stack images at 20x and 40x were acquired through the sample with and without the application of an optical clearing agent (FocusClear®). Intensity of transmitted light increased by 5-15x and islet structures unique to pancreas were clearly visualized 250-300 μm beneath the tissue surface. CBs were then placed in index matching square capillary tubes filled with FocusClear® and a standard optical clearing agent. Brightfield z-stack images were then acquired to present 3D visualization of the CB to the pathologist.