DNA Mobius self-assembling nano-architectures could lead to new nanobiooelectronics
October 5, 2010
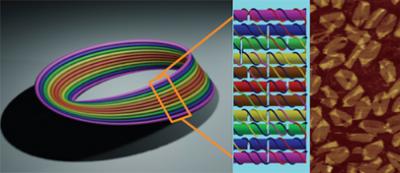
This is the design for the DNA Möbius strip. Single-stranded viral DNA is used as scaffolding and 164 short segments of DNA are used as staple strands, to create the nanostructure. The Möbius form is composed of eleven double helices, assembled in parallel (left). Each double-helical length contains a twist of 180 degrees along its central axis, before it seamlessly reconnects with itself. The central helix, (seen in red) circles around the length of the strip once. The other helices circle twice, while also twisting around the core helix by 180 degrees before reconnecting to close the Möbius loop. (Center) A small segment of the strip with the details of the helices shown. Scaffold strands are seen in blue and staple strands are different colors. To create the Möbius, 20.5 units like this were used, with the precise folding pattern pre-programmed through the design of appropriate nucleotide base-pairing. (Right) Atomic Force Microscopy image. (Nature Nanotechnology)
Scientists at the Biodesign Institute at Arizona State University’s and Department of Chemistry and Biochemistry have reproduced the shape of a Mobius strip on a remarkably tiny scale, joining up braid-like segments of DNA to create Mobius structures measuring just 50 nanometers across.
The researchers hope to eventually apply these nano-architectures to the development of biological and chemical sensing devices, nanolithography, drug delivery mechanisms and a new breed of nanoelectronics.
The team used a versatile construction method known as DNA origami and in an extension of the technique (which they refer to as DNA Kirigami), they cut the resulting Möbius shapes along their length to produce twisted ring structures and interlocking loops known as catenanes.
To form the Möbius strip in the current study, the group relied on properties of self-assembly inherent in DNA. A strand of DNA is formed from combinations of 4 nucleotide bases, adenine (A), thymine (T), cytosine (C) and guanine (G), which follow one another on the strand like necklace beads. These nucleotide beads can bind to each other according to a strict rule: A always pairs with T, C with G. Thus, a second, complementary strand of DNA binds with the first to form the DNA double helix.
In 2006, Paul Rothemund at Cal Tech demonstrated that the process of DNA self-assembly could be used to produce pre-designed 2D nanoarchitectures of astonishing variety. DNA origami emerged as a powerful tool for nanostructure design. The method relies on a long, single stranded segment of DNA, used as a structural scaffold and guided through base pairing to assume a desired shape. Short, chemically synthesized “staple strands,” composed of complementary bases are used to hold the structure in place.
After synthesis and mixing of DNA staples and scaffold strands, the structure is able to self-assemble in a single step. The technique has been used to produce remarkable nanostructures of smiley faces, squares, disks, geographic maps, and even words, at a scale of 100 nm or less. But the creation of topological forms capable of reconfiguration, like those produced by nature, has proven more challenging.
Once the tiny Möbius structures had been created, they were examined with atomic force and transmission electron microscopy. The Möbius forms displayed both right and left handed twists. Imaging permitted the handedness or chirality of each flattened nanostructure to be determined, based on the height differences observed at the overlapping areas.
Next, the team demonstrated the topological flexibility of the Möbius forms produced, using a folding and cutting—or DNA Kirigami—technique. The Möbius can be modified by cutting along the length of the strip at different locations. Cutting a Möbius along its centerline yields a new structure—a looped form containing a twist of 720 degrees or 4 half-twists. The design, which the group calls a Kirigami-Ring, is no longer a Möbius, as it has two edges and two surfaces. The Möbius may also be cut along its length one-third of the way into its width, producing a Kirigami-Catenane—a Möbius strip interlinked with a supercoiled ring.
To accurately cut the Möbius nanostructures, a technique known as strand displacement was used, in which the DNA staples holding the central helix in place are outfitted with so-called toe-hold strands which protrude from the central helix. A complementary strand binds to the toehold segment, removing the staples and allowing the Möbius to fall open into either the Kirigami-Ring or Kirigami-Catenane.
The work appears in the Oct. 4 advanced online issue of the journal Nature Nanotechnology.
More info: Arizona State University news
