Electron microscope reveals bacteria motor parts in incredible unprecedented detail
March 16, 2016

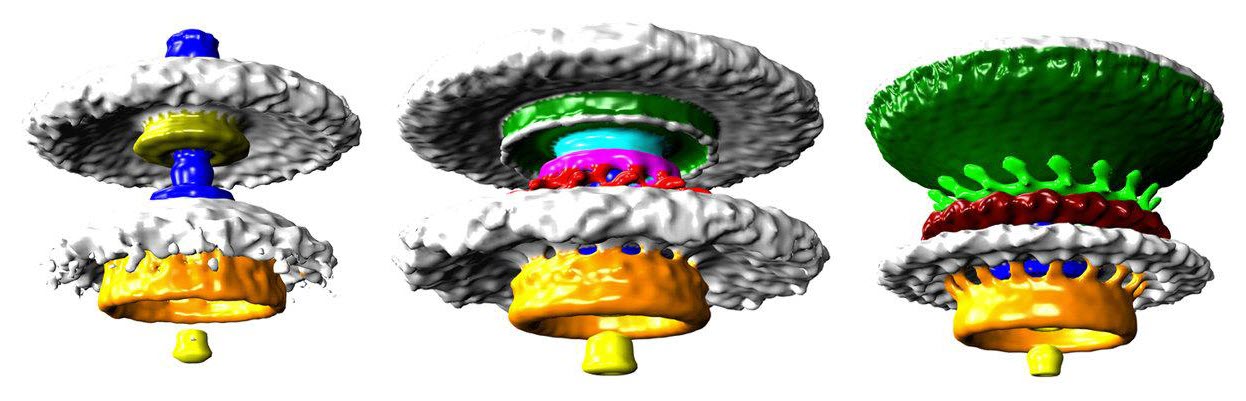
Three bacterial motors with different torque (units of torque are piconewton nanometers, pN nm) (credit: Morgan Beeby/Imperial College London)
A new study of the exotic “motors” that bacteria use to swim reveals details of how they “swim” that may make it possible to design specific drugs that sabotage the flagella (tails) in targeted bacterial species.
Using a newly installed high-powered electron microscope, researchers at Imperial College London, led by Morgan Beeby, PhD from the Department of Life Sciences, has been able visualize these motors in unprecedented detail.
They used a method called electron cryo-tomography to rapidly freeze the bacteria to -180 degrees C — preventing ice crystals from forming that would break the structure apart and allowing the researchers to image the flash-frozen motor from all angles and build up 3D models.
Their visualizations of these motors explain the differences in swimming ability, mathematically accounting for differences in motor power. The work is published in an open-access paper in Proceedings of the National Academy of Sciences.
How bacteria motors work
Bacterial flagellar motors use a clever rotational mechanism, spinning their long flagellar tail to produce a helical propeller-like motion. The team found that stronger swimmers have evolved by adding extra parts to their motors, making more powerful motors that have increased turning force, or torque.
In flagellar motors, the turning force is produced by a ring of structures called stators around the motor (similar to the structure of electric motors). The team found that Campylobacter jejuni had almost twice as many stators positioned around the motor than in Salmonella, and that these structures sat in a wider ring. More stators provide increased torque, and the increased width of the ring means individual stators exert more leverage when rotating the helical propeller.
However, not all bacteria need to be so powerful and swim through such viscous environments as stomach mucus. Instead, another bacterium the team looked at, a close relative of Vibrio cholerae, the bacterium that causes cholera, has evolved a motor with only intermediate power.
Despite motors in diverse bacteria having the same core structure, different bacteria vary widely in their swimming power. For example, Campylobacter jejuni, which causes food poisoning, can swim powerfully enough to bore through the mucus that lines the gut, an environment too thick and sticky for other bacteria to push through.
The reasons for these differences in swimming ability have remained obscure until now. By looking at distantly related bacteria from different branches of the evolutionary tree, the team speculates that the ability to alter torque in this way may have evolved up to two billion years ago.
“Entire branches of the bacterial family tree have evolved motors with different torques, leading to a diversity of species each geared to their own environment,” said Beeby. The team is now investigating how and when the evolutionary steps that altered motor torque happened.
editor’s comments: Check out the open-access paper: damn impressive engineering study! Hey, could these motor designs be genetically modified (or combined with the Drexel University method from yesterday for using electric fields to help microscopic bacteria-powered robots detect obstacles) to attack other bacteria or be sent out as scouts or on other missions? OK, just a wild and crazy idea. …
Abstract of Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold
Although it is known that diverse bacterial flagellar motors produce different torques, the mechanism underlying torque variation is unknown. To understand this difference better, we combined genetic analyses with electron cryo-tomography subtomogram averaging to determine in situ structures of flagellar motors that produce different torques, from Campylobacter and Vibrio species. For the first time, to our knowledge, our results unambiguously locate the torque-generating stator complexes and show that diverse high-torque motors use variants of an ancestrally related family of structures to scaffold incorporation of additional stator complexes at wider radii from the axial driveshaft than in the model enteric motor. We identify the protein components of these additional scaffold structures and elucidate their sequential assembly, demonstrating that they are required for stator-complex incorporation. These proteins are widespread, suggesting that different bacteria have tailored torques to specific environments by scaffolding alternative stator placement and number. Our results quantitatively account for different motor torques, complete the assignment of the locations of the major flagellar components, and provide crucial constraints for understanding mechanisms of torque generation and the evolution of multiprotein complexes.