Epigenetic changes don’t last
September 26, 2011

Changes in the epigenetic code in this Arabidopsis plant can occur spontaneously (credit: MPI for Developmental Biology)
Scientists at the Max Planck Institute of Developmental Biology in Germany have produced what they are calling the first comprehensive inventory of spontaneous epigenetic changes (genetic changes brought about by the environment), and found, contrary to previous theory, that these changes are short-lived and have minimal impact on evolution.
Using the Arabidopsis plant, the workhorse of modern plant genetics, the researchers determined how often and where in the genome epigenetic modifications occur — and how often they disappear again.
They found that epigenetic changes are many orders of magnitude more frequent than conventional DNA mutations, but often also often short-lived. They are therefore probably much less important for long-term evolution than previously thought.
The team around Detlef Weigel, director of the Department for Molecular Biology, focused on one of the most important epigenetic markers, methylation of DNA (tiny chemical building blocks, methyl groups, are attached to individual letters of the DNA, mostly to cytosines). The genetic information itself, in the form of the four different letters or nucleotides that make up the genetic code, remains unchanged in this process.
To determine the rate and distribution of methylation changes in the genome, the German biologists looked at ten Arabidopsis lines. These lines came from the same stock, but had been propagated independently for 30 generations by self-fertilization. In the genome of the last generation, the scientists then searched for differences in the methylation pattern in comparison to the common ancestor. They produced for each individual a complete map of methylated cytosines in the genome, the methylome.
“For each line, we were able to look at about 14 million cytosines,” said Claude Becker, a member of the Tübingen team. On average, every plant had almost 3 million methylated cytosines. The vast majority of these were the same in all lines, but about 6 percent had changed since the lines had become separated.
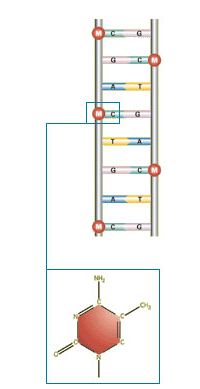
Special enzymes transfer methyl groups to the DNA cytosine building blocks. A methyl group consists of one carbon atom combined with three associated atoms of hydrogen (CH3). (Credit: Environmental Health Perspectives, 2006)
At these positions, at least one of the individuals was different, with either methylation gained or lost relative to the ancestor. Each of the lines had about 30,000 such epimutations, which was 1,000 times more than DNA mutations.
With 30,000 epimutations after 30 generations, the geneticists had expected that 1,000 epimutations occurred in each generation. When they directly compared parents and their immediate offspring, they were surprised to find that the epimutation rate was actually three to four times as high. The scientists concluded that many epimutations are apparently not stable and return to their original state after a few generations.
So it turns out that averaging the mutation rate over many generations is misleading. So Becker’s colleague Jörg Hagmann cautions not to overestimate the importance of DNA methylation during evolution: “Our experiments show that methylation changes are often reversible”. In other words: New epimutations are often not maintained over the long term.
A new epimutation must have a strong evolutionary advantage so that it can become established before being lost again. Because reverse mutations do not necessarily happen in the next generation, it is still possible that epigenetic differences contribute to inheritance of traits between parents and their children or grandparents and their grandchildren.
Another difference from ordinary mutations is that epimutations do not occur randomly, but often at the same places in the genome. While genes were often disproportionately affected, methylation of mobile DNA elements, transposons, was very stable. This appears to make sense, since it was already known that artificially induced loss of methylation has a much greater effect on the activity of transposons than of regular genes.
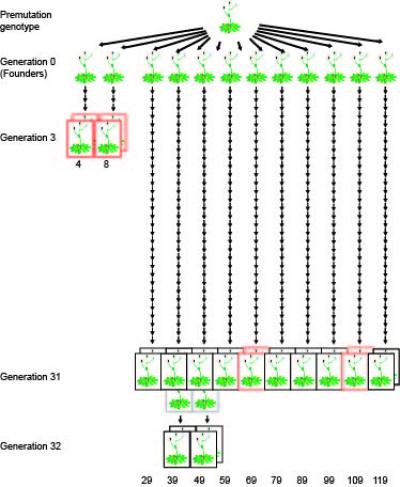
Epigenetic variations between the generations: the third generation of lines 4 and 8 serves as reference for the comparison of 10 lines after more than 30 generations (credit: MPI for Developmental Biology)
More important than the state of individual cytosines is probably the methylation of larger segments of the genome. “In each plant we found only about 30 such regions in which they differed from the other lines,” explains Becker. Hagmann adds: “Such whole-sale epigenetic changes appear to be as rare as true DNA sequence mutations”.
But these differences can appear very rapidly. The biologists discovered one region that first lost its methyl groups, only to become completely remethylated in the next generation.
What makes epigenetics interesting for human health is the fact that some epigenetic changes can be triggered by external factors. There is evidence that nutrition or the bond between children and their parents can leave traces in the genome that can be passed on to the next generation.
The limited stability of DNA methylation implies, however, that such differences do not necessarily last forever, which is probably not a bad idea, because a famine might not last forever. It also means that altered DNA methylation often cannot become subject to natural selection.
The results of the Max Planck scientists’ work demonstrate that epigenetic differences can also arise spontaneously, without drastic changes in the environment. After all, the growth conditions in the greenhouse, where each of the 10 lines was propagated, were constant.
This opens the door to further speculation. “We suspect that the epimutation increase is higher and more variable when plants grow in nature, where they are stressed all the time,” says Becker. If this were paralleled by an even higher reversion rate, the importance of epigenetics for long-term evolution would be even lower.
Ref.: Claude Becker, et al., Spontaneous epigenetic variation in the Arabidopsis thaliana methylome, Nature, 2011; [DOI: 10.1038/nature10555]