First complete pictures of cells’ DNA-copying machinery
November 3, 2015
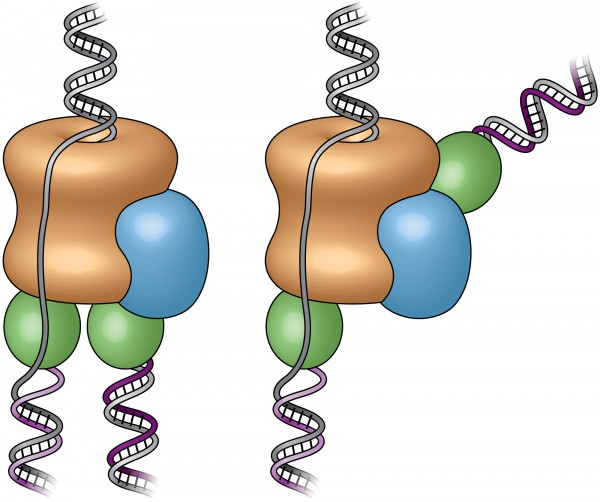
These cartoons show the old “textbook” view of the replisome, left, and the new view, right, revealed by electron micrograph images in the current study. Prior to this study, scientists believed the two polymerases (green) were located at the bottom (or back end) of the helicase (tan), adding complementary DNA strands to the split DNA to produce copies side by side. The new images reveal that one of the polymerases is actually located at the front end (top) of the helicase. The scientists are conducting additional studies to explore the biological significance of this unexpected location. (credit: Brookhaven National Laboratory)
The first-ever electron microscope images of the protein complex that unwinds, splits, and copies double-stranded DNA reveal something rather different from the standard textbook view.
The images, created by scientists at the U.S. Department of Energy’s Brookhaven National Laboratory with partners from Stony Brook University and Rockefeller University, offer new insight into how this molecular machinery functions, including new possibilities about its role in DNA “quality control” and cell differentiation.
Huilin Li, a biologist with a joint appointment at Brookhaven Lab and Stony Brook University says the new images show the fully assembled and fully activated helicase protein complex — which encircles and separates the two strands of the DNA double helix as it passes through a central pore in the structure — and how the helicase coordinates with the two polymerase enzymes that duplicate each strand to copy the genome.
Three blind men and an elephant
Studying this molecular machinery, known collectively as a “replisome,” and the details of its DNA-copying process can help scientists understand what happens when DNA is miscopied — a major source of mutation that can lead to cancer. Scientists can also learn more about how a single cell can eventually develop into the many cell types that make up a multicellular organism.
“All the textbook drawings and descriptions of how a replisome should look and work are based on biochemical and genetic studies,” Li said, likening the situation to the famous parable of the three blind men trying to describe an elephant, each looking at only one part.
To test these assumptions, Li’s group turned to electron microscopy (EM). The team’s first-ever images of an intact replisome revealed that only one of the polymerases is located at the back of the helicase.
The other is on the front side of the helicase, where the helicase first encounters the double-stranded helix. This means that while one of the two split DNA strands is acted on by the polymerase at the back end, the other has to thread itself back through or around the helicase to reach the front-side polymerase before having its new complementary strand assembled.
Unforeseen functions?
The counterintuitive position of one polymerase at the front of the helicase suggests that it may have an unforeseen function. The authors suggest several possibilities, including keeping the two “daughter” strands separate to help organize them during replication and cell division. It might also be possible that, as the single strand moves over other portions of the structure, some “surveillance” protein components check for lesions or mistakes in the nucleotide sequence before it gets copied — a sort of molecular quality control.
This architecture could also potentially play an important role in developmental biology by providing a pathway for treating the two daughter strands differently. Many modifications to DNA, including how it is packaged with other proteins, control which of the many genes in the sequence are eventually expressed in cells. An asymmetric replisome may result in asymmetric treatment of the two daughter strands during cell division, an essential step for making different tissues within a multicellular organism.
“Clearly, further studies will be required to understand the functional implications of the unexpected replisome architecture reported here,” concludes the researchers’ paper published Monday (Nov. 2) online by the journal Nature Structural & Molecular Biology.
Brookhaven National Laboratory | Three-dimensional structure of the active DNA helicase bound to the front-end DNA polymerase (Pol epsilon). The DNA polymerase epsilon (green) sits on top rather than the bottom of the helicase.
Abstract of The Architecture of a Eukaryotic Replisome
At the eukaryotic DNA replication fork, it is widely believed that the Cdc45–Mcm2–7–GINS (CMG) helicase is positioned in front to unwind DNA and that DNA polymerases trail behind the helicase. Here we used single-particle EM to directly image a Saccharomyces cerevisiae replisome. Contrary to expectations, the leading strand Pol ε is positioned ahead of CMG helicase, whereas Ctf4 and the lagging-strand polymerase (Pol) α–primase are behind the helicase. This unexpected architecture indicates that the leading-strand DNA travels a long distance before reaching Pol ε, first threading through the Mcm2–7 ring and then making a U-turn at the bottom and reaching Pol ε at the top of CMG. Our work reveals an unexpected configuration of the eukaryotic replisome, suggests possible reasons for this architecture and provides a basis for further structural and biochemical replisome studies.