First genetic-based tool to detect circulating cancer cells in blood — lights up cancer cells
November 20, 2014
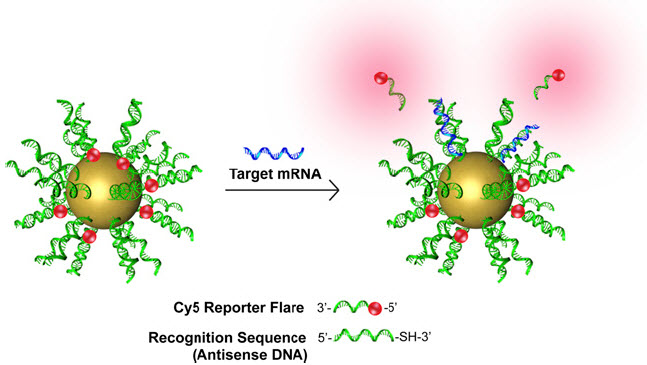
NanoFlares light up (red clouds) individual cells if a cancer (in this study, breast cancer) biomarker (messenger RNA, blue) is detected by recognition DNA (green) molecules coated on gold nanospheres and containing a fluorescent chemical (red) reporter flare (credit: Tiffany L. Halo et al./PNAS)
Northwestern University scientists have demonstrated a simple but powerful tool called NanoFlare that can detect live cancer cells in the bloodstream, potentially long before settling somewhere in the body and forming a dangerous tumor.
The NanoFlare technology is the first genetic-based approach that is able to detect live circulating tumor cells out of the complex matrix that is human blood — no easy feat. The NanoFlares are tiny spherical nucleic acids with gold nanoparticle cores outfitted with single-stranded DNA “flares” (glowing markers).
In a breast cancer study, the NanoFlares easily entered cells and lit up the cell if a biomarker target was present, even if only a trace amount.
“This technology has the potential to profoundly change the way breast cancer in particular and cancers in general are both studied and treated,” said Chad A. Mirkin, PhD, a noted nanomedicine expert and a corresponding author of the study.
Mirkin’s colleagues C. Shad Thaxton, M.D. and Dr. Chonghui Cheng, M.D., both of Northwestern University Feinberg School of Medicine, are also corresponding authors.
The research team, in a paper published the week of Nov. 17 in the Proceedings of the National Academy of Sciences (PNAS), reports two key innovations:
- The ability to track tumor cells in the bloodstream based on genetic content located within the cell itself, as opposed to using proteins located on the cell’s surface (current technology)
- The ability to collect the cells in live form, so they may be studied and used to inform researchers and clinicians as to how to treat a disease — an important step toward personalized medicine
“Cancers are very genetically diverse, and it’s important to know what cancer subtype a patient has,” Mirkin said. “Now you can think about collecting a patient’s cells and studying how those cells respond to different therapies. The way a patient responds to treatment depends on the genetic makeup of the cancer.”
Mirkin is the George B. Rathmann Professor of Chemistry in the Weinberg College of Arts and Sciences and professor of medicine, chemical and biological engineering, biomedical engineering and materials science and engineering.
How it works
A NanoFlare is designed to recognize a specific genetic code snippet associated with a cancer. The core nanoparticle, only 13 nanometers in diameter, enters cells, and the NanoFlare seeks its target. If the genetic target is present in the cell, the NanoFlare binds to it and the reporter “flare” is released to produce a fluorescent signal. The researchers then can isolate those cells.
“The NanoFlare turns on a light in the cancer cells you are looking for,” said Thaxton, an assistant professor of urology at Feinberg. “That the NanoFlares are effective in the complex matrix of human blood is a great technical advance. We can find small numbers of cancer cells in blood, which really is like searching for a needle in a haystack.”
Once they identified the cancer cells, the researchers were able to separate them from normal cells. This ability to isolate, culture and grow the cancer cells will allow researchers to zero in on the cancer cells that matter to the health of the patient. Most circulating tumor cells may not metastasize, and analysis of the cancer cells could identify those that will.
“This could lead to personalized therapy where we can look at how an individual’s cells respond to different therapeutic cocktails,” said Mirkin, whose lab developed NanoFlares in 2007.
The study
In the study, the genetic targets were messenger RNA (mRNA) that code for certain proteins known to be biomarkers for aggressive breast cancer cells.
The research team first used the blood of healthy individuals, spiking some of the blood with living breast cancer cells to see if the NanoFlares could detect them. (Unspiked blood was used as a control.)
Cheng, an assistant professor of medicine in hematology/oncology at Feinberg, provided the cell lines and NanoFlare targets that the researchers used to model blood samples taken from breast cancer patients.
The research team tested four different NanoFlares, each with a different genetic target relevant to breast cancer metastasis. The technology successfully detected the cancer cells with less than 1 percent incidence of false-negative results.
Currently, in another study, the researchers are focused on detecting circulating tumor cells in the blood of patients with a diagnosis of breast cancer.
“When it comes to detecting and treating cancer, the mantra is the earlier, the better,” Thaxton said. “This technology may enable us to better detect circulating cancer cells and provides another tool to add to the toolkit of cancer diagnosis.”
Mirkin, Thaxton and Cheng are members of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University.
The National Cancer Institute, the National Institutes of Health, the American Cancer Society, the Air Force Office of Scientific Research, and the Howard Hughes Medical Institute supported the research.
Abstract of NanoFlares for the detection, isolation, and culture of live tumor cells from human blood
Metastasis portends a poor prognosis for cancer patients. Primary tumor cells disseminate through the bloodstream before the appearance of detectable metastatic lesions. The analysis of cancer cells in blood—so-called circulating tumor cells (CTCs)—may provide unprecedented opportunities for metastatic risk assessment and investigation. NanoFlares are nanoconstructs that enable live-cell detection of intracellular mRNA. NanoFlares, when coupled with flow cytometry, can be used to fluorescently detect genetic markers of CTCs in the context of whole blood. They allow one to detect as few as 100 live cancer cells per mL of blood and subsequently culture those cells. This technique can also be used to detect CTCs in a murine model of metastatic breast cancer. As such, NanoFlares provide, to our knowledge, the first genetic-based approach for detecting, isolating, and characterizing live cancer cells from blood and may provide new opportunities for cancer diagnosis, prognosis, and personalized therapy.