How to capture videos of brains in real time
June 28, 2017
A team of scientists has peered into a mouse brain with light, capturing live neural activity of hundreds of individual neurons in a 3D section of tissue at video speed (30 Hz) in a single recording for the first time.
Besides serving as a powerful research tool, this discovery means it may now be possible to “alter stimuli in real time based on what we see going on in the animal’s brain,” said Rockefeller University’s Alipasha Vaziri, senior author of an open-access paper published June 26, 2017 in Nature Methods.
By dramatically reducing the time and computational resources required to generate such an image, the algorithm opens the door to more sophisticated experiments, says Vaziri, head of the Rockefeller Laboratory of Neurotechnology and Biophysics. “Our goal is to better understand brain function by monitoring the dynamics within densely interconnected, three-dimensional networks of neurons,” Vaziri explained.
The research “may open the door to a range of applications, including real-time whole-brain recording and closed-loop interrogation of neuronal population activity in combination with optogenetics and behavior,” the paper authors suggest.
Watching mice think in real time
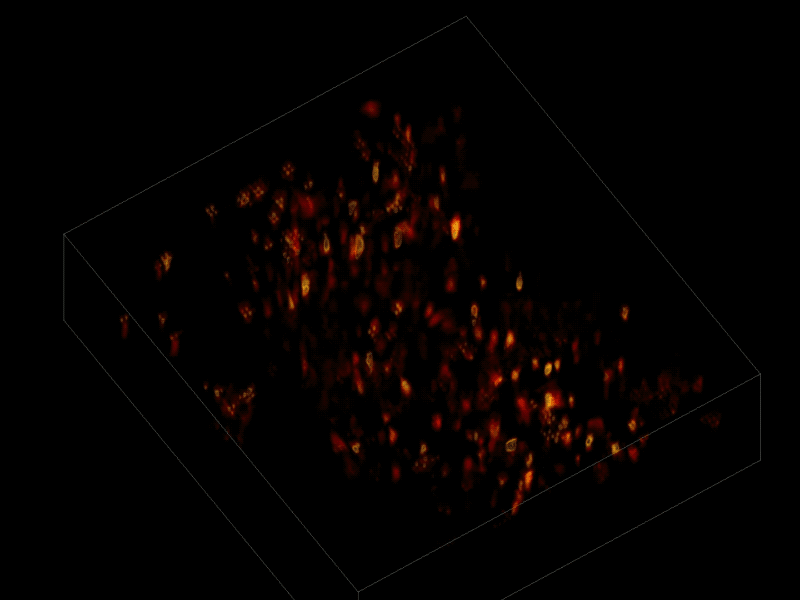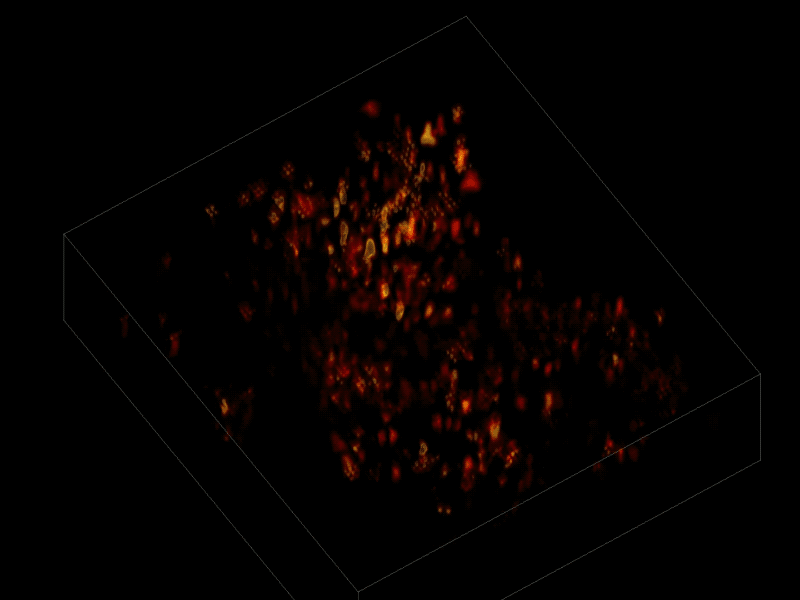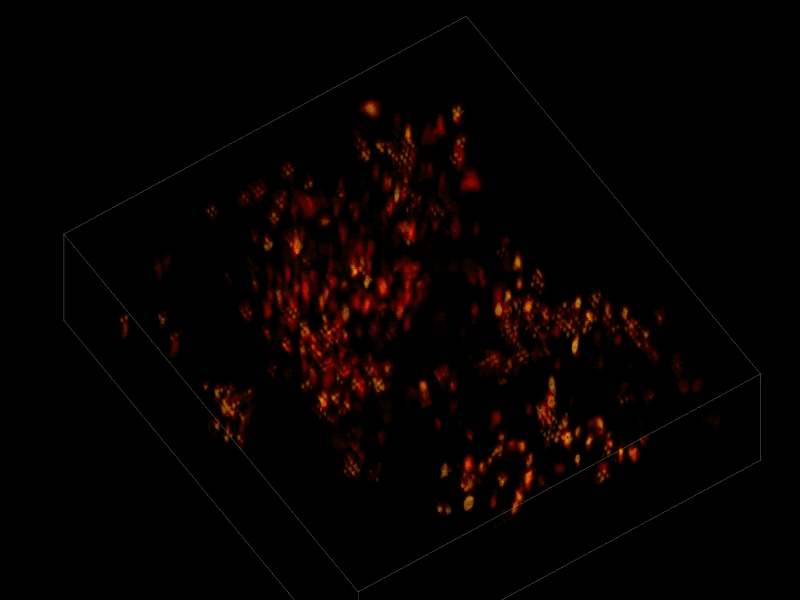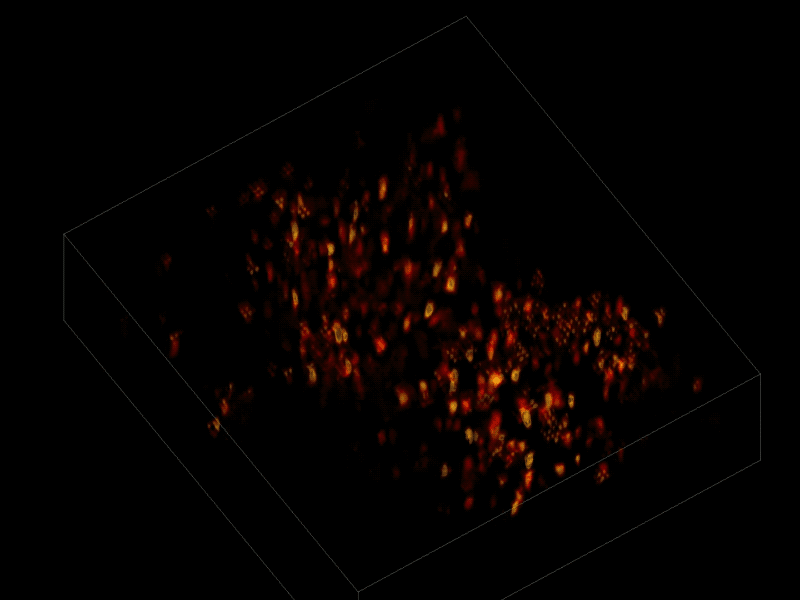
The scientists first engineered the animals’ neurons to fluoresce (glow), using a method called optogenetics. The stronger the neural signal, the brighter the cells shine. To capture this activity, they used a technique known as “light-field microscopy,” in which an array of lenses generates views from a variety of perspectives. These images are then combined to create a three-dimensional rendering, using a new algorithm called “seeded iterative demixing” (SID) developed by the team.
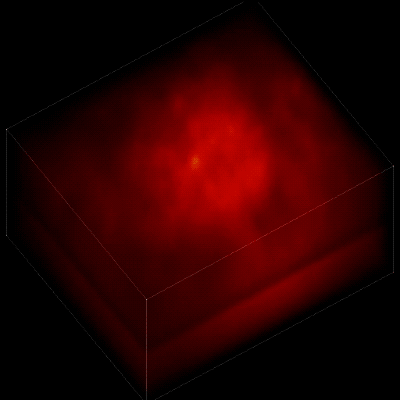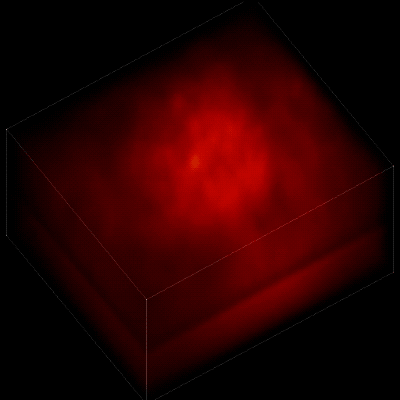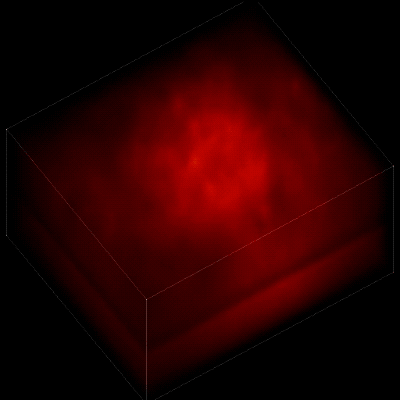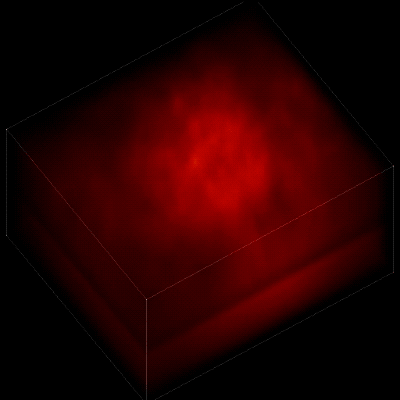
Without the new algorithm, the individual neurons are difficult to distinguish. (credit: The Rockefeller University)
To record the activity of all neurons at the same time, their images have to be captured on a camera simultaneously. In earlier research, this has made it difficult to distinguish the signals emitted by all cells as the light from the mouse’s neurons bounces off the surrounding, opaque tissue. The neurons typically show up as an indistinct, flickering mass.
The SID algorithm now makes it possible to simultaneously capture both the location of the individual neurons and the timing of their signals within a three-dimensional section of brain containing multiple layers of neurons, down to a depth of 0.38 millimeters.* Vaziri and his colleagues were able to track the precise coordinates of hundreds of active neurons over an extended period of time in mice that were awake and had the option of walking on a customized treadmill.
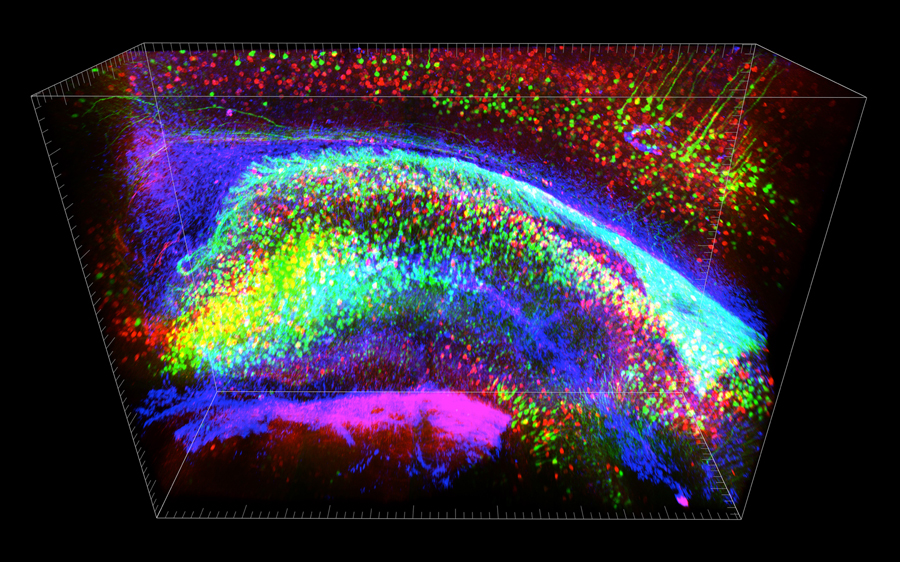
Three-dimensional view of stained hippocampus with Stanford University’s CLARITY system, showing fluorescent-expressing neurons (green), connecting interneurons (red) and supporting glia (blue). (Credit: Deisseroth lab)
Researchers were previously only able to look into brains of transparent organisms, such as the larvae of zebrafish. Stanford University scientists were able to image mouse brains in 3D (with the CLARITY system), but only for static images.
* “SID can capture neuronal dynamics in vivo within a volume of 900 × 900 × 260 μm located as deep as 380 μm in the mouse cortex or hippocampus at a 30-Hz volume rate while discriminating signals from neurons as close as 20 μm apart, at a computational cost three orders of magnitude less than that of frame-by-frame image reconstruction.” — Tobias Nöbauer et al./Nature Methods
UPDATE June 29, 2017 — Added: “The research ‘may open the door to a range of applications, including real-time whole-brain recording and closed-loop interrogation of neuronal population activity in combination with optogenetics and behavior,’ the paper authors suggest.”
Abstract of Video rate volumetric Ca2+ imaging across cortex using seeded iterative demixing (SID) microscopy
Light-field microscopy (LFM) is a scalable approach for volumetric Ca2+ imaging with high volumetric acquisition rates (up to 100 Hz). Although the technology has enabled whole-brain Ca2+ imaging in semi-transparent specimens, tissue scattering has limited its application in the rodent brain. We introduce seeded iterative demixing (SID), a computational source-extraction technique that extends LFM to the mammalian cortex. SID can capture neuronal dynamics in vivo within a volume of 900 × 900 × 260 μm located as deep as 380 μm in the mouse cortex or hippocampus at a 30-Hz volume rate while discriminating signals from neurons as close as 20 μm apart, at a computational cost three orders of magnitude less than that of frame-by-frame image reconstruction. We expect that the simplicity and scalability of LFM, coupled with the performance of SID, will open up a range of applications including closed-loop experiments.