New method enables discovery of 3D structures for molecules important to medicine
February 19, 2016

If you zap a crystal (green, left) containing highly ordered protein molecules with X-rays, the X-rays scatter and produce useful regular patterns of spots known as Bragg peaks (red dots). But if the protein in the crystal is less ordered or disordered (right), the X-rays produce some spots along with patterns of light and shade known as a continuous diffraction pattern that’s not useful. (credit: Eberhard Reimann/DESY)
Researchers have overcome a long-standing technical barrier to imaging 3D structures of thousands of molecules important to medicine and biology.
The 3D structures of many protein molecules have been discovered using a technique called X-ray crystallography, but the method relies on scientists being able to produce highly ordered crystals containing the protein molecules in a regular arrangement. When X-rays are shone on highly ordered crystals, the X-rays scatter and produce regular patterns of spots called Bragg peaks (see figure above, left). High-quality Bragg peaks contain the information to produce high-resolution 3D structures of proteins.
Unfortunately, many important and complex biomolecules do not form highly ordered crystals; instead, the protein arrangements are slightly disordered. When X-rays are shone on these more disordered crystals, a smaller number of Bragg peaks are produced, along with a vague pattern of light and shadow known as a continuous diffraction pattern (right).
In the past, scientists discarded these less-than-perfect crystals. Unfortunately, many of the molecules forming disordered crystals are important molecular complexes such as those that span cell membranes.
X-raying crystal patterns to detect hidden protein structures
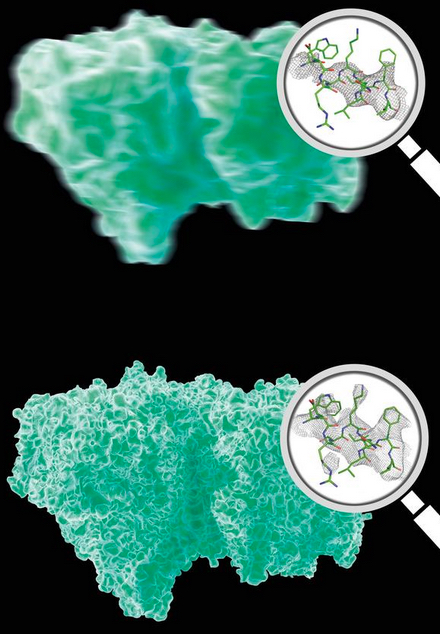
Analysis of Bragg peaks alone (top) reveals far less details than analysis of the high-res continuous diffraction pattern (bottom). Magnifying glasses show actual data. (credit: DESY, Eberhard Reimann)
So a team led by Professor Henry Chapman from the Center for Free-Electron Laser Science at DESY in Hamburg, Germany turned to the world’s most powerful X-ray laser: the SLAC LCLS at Stanford University.
Kartik Ayyer, PhD., lead author of the article in Nature, explains that the method uses an approach similar to that used to image a single molecule.
“If you would shoot X-rays on a single molecule, it would produce a continuous diffraction pattern free of any Bragg spots,” he says. “The pattern would be extremely weak, however, and very difficult to measure. But the ‘background’ in our crystal analysis is like accumulating many shots from individually aligned single molecules. We essentially just use the crystal as a way to get a lot of single molecules, aligned in common orientations, into the beam.”
As the model protein, the researchers crystallized photosystem II (PSII), a large membrane–protein complex of photosynthesis that plants use to produce oxygen for life on Earth.
After exposing the crystal to X-rays, the researchers first analyzed the Bragg peaks of PSII to produce a low-resolution outline of the 3D structure (figure above, top). They then improved this data, using an algorithm, to analyze the continuous diffraction pattern and produced a higher-resolution 3D structure (figure, bottom).
This novel method means that imperfect crystals containing a slightly disordered protein arrangement can now be used to “directly view large protein complexes in atomic detail,” says Chapman. “This kind of continuous diffraction has actually been seen for a long time from many different poorly diffracting crystals,” says Chapman. “It wasn’t understood that you can get structural information from it and so analysis techniques suppressed it.
“We’re going to be busy to see if we can solve [additional] structures of molecules from old discarded data.”
Abstract of Macromolecular diffractive imaging using imperfect crystals
The three-dimensional structures of macromolecules and their complexes are mainly elucidated by X-ray protein crystallography. A major limitation of this method is access to high-quality crystals, which is necessary to ensure X-ray diffraction extends to sufficiently large scattering angles and hence yields information of sufficiently high resolution with which to solve the crystal structure. The observation that crystals with reduced unit-cell volumes and tighter macromolecular packing often produce higher-resolution Bragg peaks suggests that crystallographic resolution for some macromolecules may be limited not by their heterogeneity, but by a deviation of strict positional ordering of the crystalline lattice. Such displacements of molecules from the ideal lattice give rise to a continuous diffraction pattern that is equal to the incoherent sum of diffraction from rigid individual molecular complexes aligned along several discrete crystallographic orientations and that, consequently, contains more information than Bragg peaks alone. Although such continuous diffraction patterns have long been observed—and are of interest as a source of information about the dynamics of proteins—they have not been used for structure determination. Here we show for crystals of the integral membrane protein complex photosystem II that lattice disorder increases the information content and the resolution of the diffraction pattern well beyond the 4.5-ångström limit of measurable Bragg peaks, which allows us to phase the pattern directly. Using the molecular envelope conventionally determined at 4.5 ångströms as a constraint, we obtain a static image of the photosystem II dimer at a resolution of 3.5 ångströms. This result shows that continuous diffraction can be used to overcome what have long been supposed to be the resolution limits of macromolecular crystallography, using a method that exploits commonly encountered imperfect crystals and enables model-free phasing.