New synthetic gene circuits can perform complex bio-logic tasks
September 11, 2014
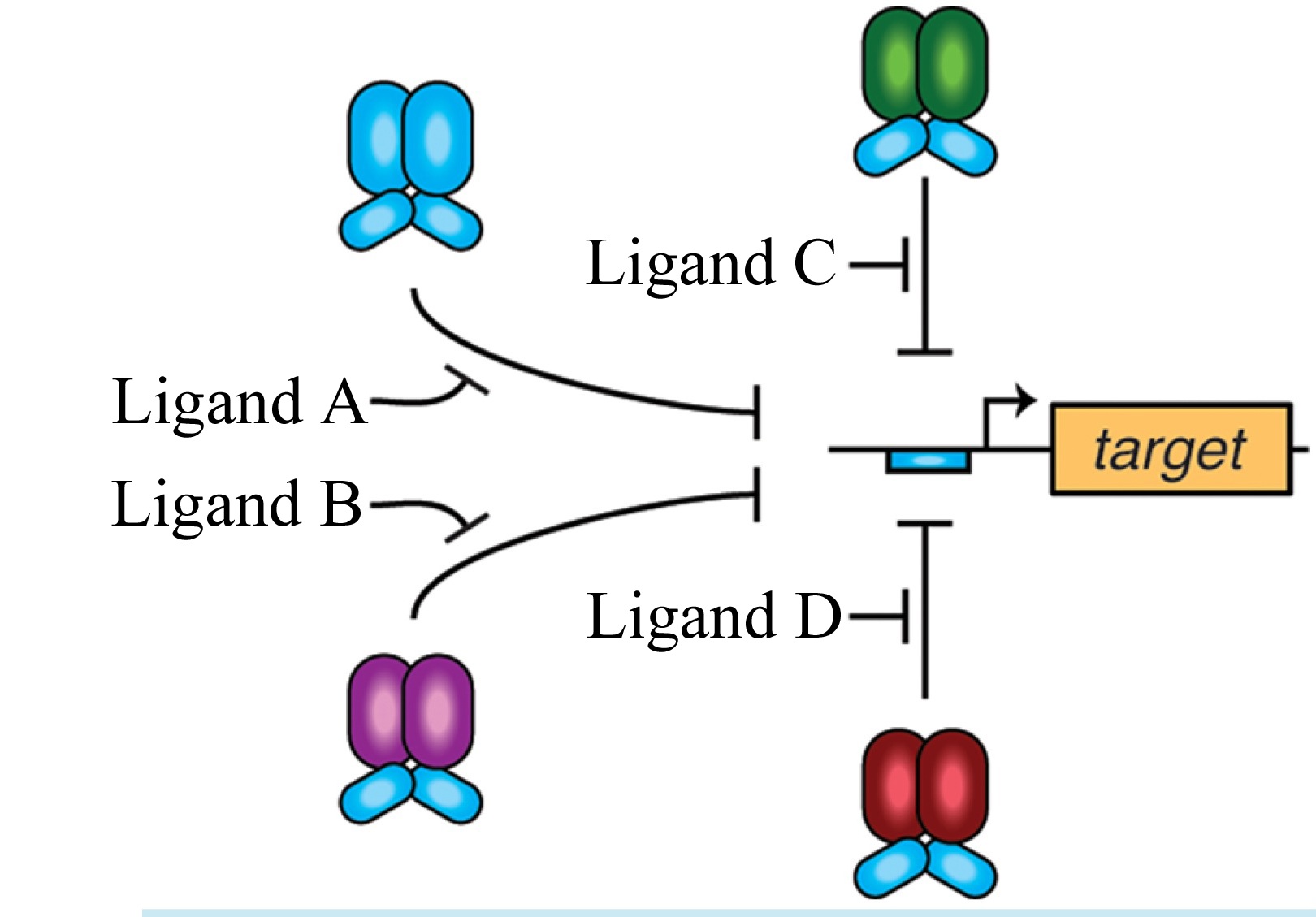
Scientists at Rice University and the University of Kansas Medical Center are using multiple chimeric transcription factors as logic circuits to perform complex tasks in cells. The circuits are triggered when modular protein domains sense the presence of specific chemical combinations in a cell. (Credit: Bennett Lab/Rice University)
Researchers at Rice University and the University of Kansas Medical Center are making genetic circuits that can perform complex tasks by swapping protein building blocks.
The modular genetic circuits, which are engineered from parts of otherwise unrelated bacterial genomes, can be set up to handle multiple chemical inputs simultaneously with a minimum of interference from their neighbors.
The work, reported in the American Chemical Society journal ACS Synthetic Biology, gives scientists more options as they design synthetic cells for specific tasks, such as production of biofuels, environmental remediation, or treatments for human diseases.
The researchers are creating complex genetic logic circuits similar to those used to build traditional computers and electrical devices. In a simple circuit, if one input and another input are both present (AND gate), the circuit carries out its instruction. With genetic circuitry based on this type of Boolean logic, a genetic logic circuit might prompt the creation of a specific protein when it senses two chemicals — or prompt a cell’s DNA to repress the creation of that protein.
Simple circuits have become easier to create as synthetic biologists develop more tools, but they require more sophisticated tools for complex problems. Rice’s Matthew Bennett and his colleagues are intent upon following a path similar to that of computer programmers, whose capabilities grew from simple Pong to the immersive worlds of modern games.
Programming cells
“One of the ultimate goals of this technology is to allow cells to sense and respond to their environment in programmatic ways,” said Bennett, an assistant professor of biochemistry and cell biology. “We want to be able to program cells to go into an environment and do what they’re supposed to do.
“Right now, one of the main ways we do that is through transcriptional logic gates. These are akin to electronic circuits — the logic gates in our computers. In cells, they work a little bit differently, but there are a lot of parallels.”
Logic gates designed by Bennett’s team and others react in a programmed way when they sense chemicals in their immediate environment. If certain combinations of chemicals are present in the environment, the gate will turn on a gene that may either repress or promote the expression of a protein.
The research, led by Rice graduate student David Shis, drew from a genetic toolbox of chimeric (with parts from different sources) transcription factors. These modular proteins incorporate the gene regulatory capacity of one transcription factor and the environmental sensing capabilities of another.
The researchers demonstrated that as many as four chimeras with the same DNA-binding modules can work together and serve as gates with multiple inputs, either repressing — or overriding the repression of — specific genes. They successfully tested the ability of chimera combinations in the bacteria Escherichia coli to up- or down-regulate the expression of a gene encoding green fluorescent protein.
“Often, when you make a genetic logic gate, you have to have many genes in the background to allow the gate to work,” Bennett said. “We’ve been able to eliminate the need for that by programming transcription factors — which are specific proteins that turn genes on and off — to respond to their environment directly and activate a specific gene in a very modular way.
“We can now program both environmental sensing and downstream genetic regulation into the same module,” he said.
Applications
Bennett sees synthetic biology addressing many issues. “We might be able to use cells to report on, or remediate, environmental pollution. Or we might be able to program them to find a tumor in your body and respond to it. To do that, we need to be able to instruct cells to sense the environment of the tumor and, depending on what chemicals the cells detect, respond accordingly.”
Metabolic engineers might find complex synthetic circuits that are able to adjust on the fly, he said. “In fermentation, for example, you might want gene regulation in the cells to change as a process evolves. These new circuits can sense different sugars in the culture and direct gene regulation to maximize production.”
The National Institutes of Health, through the joint National Science Foundation/National Institute of General Medical Sciences Mathematical Biology Program, and the Robert A. Welch Foundation supported the research.
Abstract of ACS Synthetic Biology paper
In prokaryotes, the construction of synthetic, multi-input promoters is constrained by the number of transcription factors that can simultaneously regulate a single promoter. This fundamental engineering constraint is an obstacle to synthetic biologists because it limits the computational capacity of engineered gene circuits. Here, we demonstrate that complex multi-input transcriptional logic gating can be achieved through the use of ligand-inducible chimeric transcription factors assembled from the LacI/GalR family. These modular chimeras each contain a ligand-binding domain and a DNA-binding domain, both of which are chosen from a library of possibilities. When two or more chimeras have the same DNA-binding domain, they independently and simultaneously regulate any promoter containing the appropriate operator site. In this manner, simple transcriptional AND gating is possible through the combination of two chimeras, and multiple-input AND gating is possible with the simultaneous use of three or even four chimeras. Furthermore, we demonstrate that orthogonal DNA-binding domains and their cognate operators allow the coexpression of multiple, orthogonal AND gates. Altogether, this work provides synthetic biologists with novel, ligand-inducible logic gates and greatly expands the possibilities for engineering complex synthetic gene circuits.