Smart robot accelerates cancer treatment research by finding optimal treatment combinations
September 23, 2015
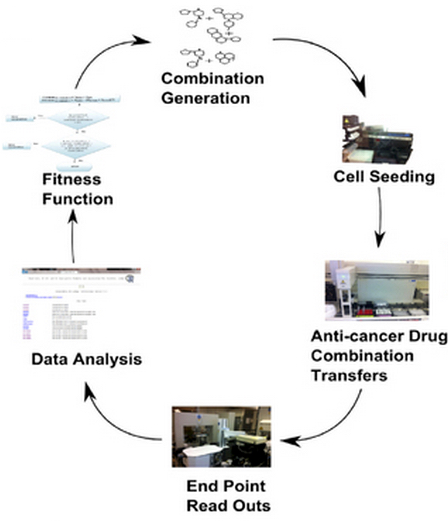
Iterative search for anti-cancer drug combinations. The procedure starts by generating an initial generation (population) of drug combinations randomly or guided by biological prior knowledge and assumptions. In each iteration the aim is to propose a new generation of drug combinations based on the results obtained so far. The procedure iterates through a number of generations until a stop criterion for a predefined fitness function is satisfied. (credit: M. Kashif et al./Scientific Reports)
A new smart research system developed at Uppsala University accelerates research on cancer treatments by finding optimal treatment drug combinations. It was developed by a research group led by Mats Gustafsson, Professor of Medical Bioinformatics.
The “lab robot” system plans and conducts experiments with many substances, and draws its own conclusions from the results. The idea is to gradually refine combinations of substances so that they kill cancer cells without harming healthy cells.
Instead of just combining a couple of substances at a time, the new lab robot can handle about a dozen drugs simultaneously. The future aim is to handle many more, preferably hundreds.
There are a few such laboratories in the world with this type of lab robot, but researchers “have only used the systems to look for combinations that kill the cancer cells, not taking the side effects into account,” says Gustafsson.
The next step: make the robot system more automated and smarter. The scientists also want to build more knowledge into the guiding algorithm of the robot, such as prior knowledge about drug targets and disease pathways.
For patients with the same cancer type returning multiple times, sometimes the cancer cells develop resistance against the pharmacotherapy used. The new robot systems may also become important in the efforts to find new drug compounds that make these resistant cells sensitive again.
The research is described in an open-access article published Tuesday (Sept. 22, 2015) in Scientific Reports.
Abstract of In vitro discovery of promising anti-cancer drug combinations using iterative maximisation of a therapeutic index
In vitro-based search for promising anti-cancer drug combinations may provide important leads to improved cancer therapies. Currently there are no integrated computational-experimental methods specifically designed to search for combinations, maximizing a predefined therapeutic index (TI) defined in terms of appropriate model systems. Here, such a pipeline is presented allowing the search for optimal combinations among an arbitrary number of drugs while also taking experimental variability into account. The TI optimized is the cytotoxicity difference (in vitro) between a target model and an adverse side effect model. Focusing on colorectal carcinoma (CRC), the pipeline provided several combinations that are effective in six different CRC models with limited cytotoxicity in normal cell models. Herein we describe the identification of the combination (Trichostatin A, Afungin, 17-AAG) and present results from subsequent characterisations, including efficacy in primary cultures of tumour cells from CRC patients. We hypothesize that its effect derives from potentiation of the proteotoxic action of 17-AAG by Trichostatin A and Afungin. The discovered drug combinations against CRC are significant findings themselves and also indicate that the proposed strategy has great potential for suggesting drug combination treatments suitable for other cancer types as well as for other complex diseases.