Software equal to or better than humans at cataloging published science data
December 2, 2014
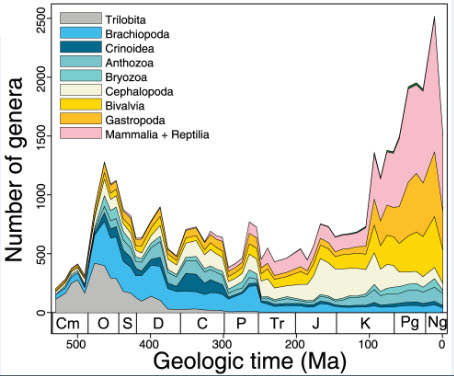
Computer-generated genus-level diversity curves (credit: Shanan E. Peters et al./PLOS ONE)
A computer system called PaleoDeepDive has equaled (or bested in some categories) scientists at the complex task of extracting data from scientific publications and placing it in a database that catalogs the results of tens of thousands of individual studies.
The development, described in the current issue of PLoS, marks a milestone in the quest to rapidly and precisely summarize, collate, and index the vast output of scientists around the globe, says first author Shanan Peters, a professor of geoscience at UW-Madison.
Peters and colleagues set up the face-off between their new machine reading system and the human scientists who had manually entered data into the Paleobiology Database. This repository, compiled by hundreds of researchers, is the destination for data from paleontology studies funded by the National Science Foundation and other agencies internationally.
A probabilistic approach
The knowledge produced by paleontologists is fragmented into hundreds of thousands of publications. Yet many research questions require what Peters calls a “synthetic approach: For example, how many species were on the planet at any given time?”
PaleoDeepDive mimics the human activities needed to assemble the Paleobiology Database. “We extracted the same data from the same documents and put it into the exact same structure as the human researchers, allowing us to rigorously evaluate the quality of our system, and the humans,” Peters says.
Instead of trying to divine the single correct meaning, the tactic was to “to look at the entire problem of extraction as a probabilistic problem,” says Christopher Ré, who guided the software development while a UW professor of computer sciences.
Computers often have trouble deciphering even simple-sounding statements, Ré says. Ré imagines a study containing the terms “Tyrannosaurus rex” and “Alberta, Canada.” Is Alberta where the fossil was found, or where it is stored? “We take a more relaxed approach: There is some chance that these two are related in this manner, and some chance they are related in that manner.”
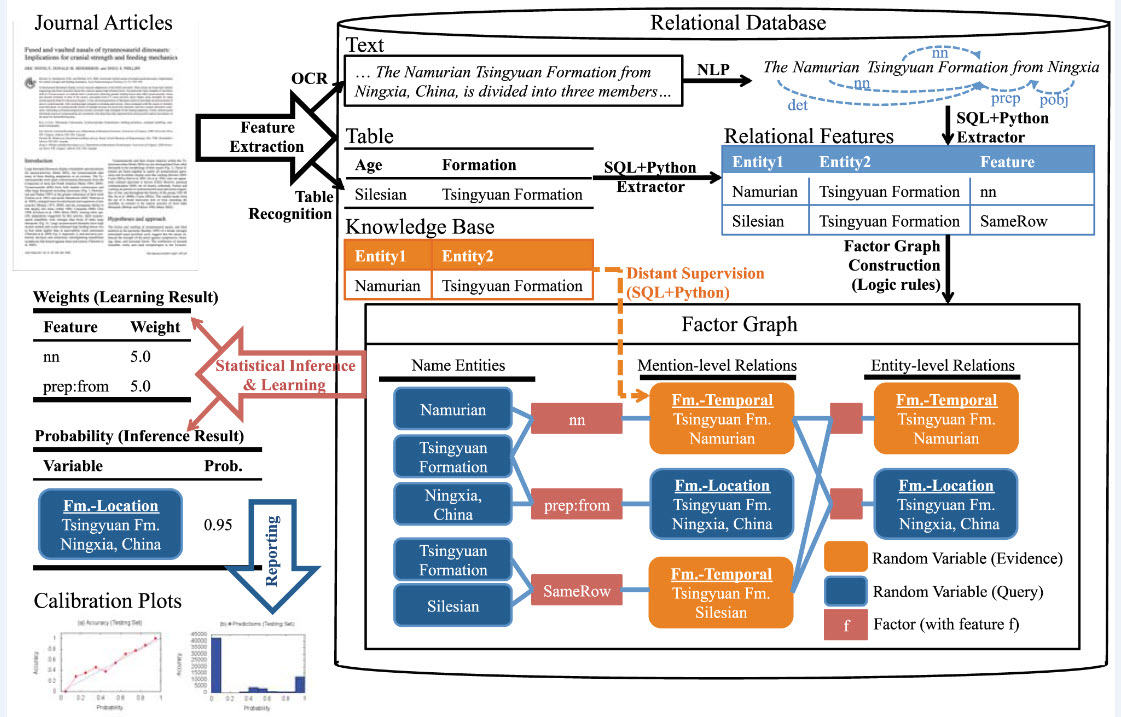
Schematic representation of the PaleoDeepDive workflow (credit: Shanan E. Peters et al./PLOS ONE)
In these large-data tasks, PaleoDeepDive has a major advantage, Peters says. “Information that was manually entered into the Paleobiology Database by humans cannot be assessed or enhanced without going back to the library and re-examining original documents. Our machine system, on the other hand, can extend and improve results essentially on the fly as new information is added.”
Further advantages can result from improvements in the computer tools. “As we get more feedback and data, it will do a better job across the board.”
The project required a million hours of computer time. It also required access to tens of thousands of articles, says Jacquelyn Crinion, assistant director of licensing and acquisitions services at the UW–Madison General Library System. And the download volume threatened logjams in document delivery. Eventually, Elsevier gave the UW-Madison team broad access to 10,000 downloads per week.
As text- and data-mining takes off, Crinion says the library system and publishers will adapt. “The challenge for all of us is to provide specialized services for researchers while continuing to meet the core needs of the vast majority of our customers.”
The Paleobiology Database has already generated hundreds of studies about the history of life, Peters says. “Ultimately, we hope to have the ability to create a computer system that can do almost immediately what many geologists and paleontologists try to do on a smaller scale over a lifetime: read a bunch of papers, arrange a bunch of facts, and relate them to one another in order to address big questions.”
Abstract of A Machine Reading System for Assembling Synthetic Paleontological Databases
Many aspects of macroevolutionary theory and our understanding of biotic responses to global environmental change derive from literature-based compilations of paleontological data. Existing manually assembled databases are, however, incomplete and difficult to assess and enhance with new data types. Here, we develop and validate the quality of a machine reading system, PaleoDeepDive, that automatically locates and extracts data from heterogeneous text, tables, and figures in publications. PaleoDeepDive performs comparably to humans in several complex data extraction and inference tasks and generates congruent synthetic results that describe the geological history of taxonomic diversity and genus-level rates of origination and extinction. Unlike traditional databases, PaleoDeepDive produces a probabilistic database that systematically improves as information is added. We show that the system can readily accommodate sophisticated data types, such as morphological data in biological illustrations and associated textual descriptions. Our machine reading approach to scientific data integration and synthesis brings within reach many questions that are currently underdetermined and does so in ways that may stimulate entirely new modes of inquiry.