Structure of key control element behind protein misfolding identified
March 1, 2012
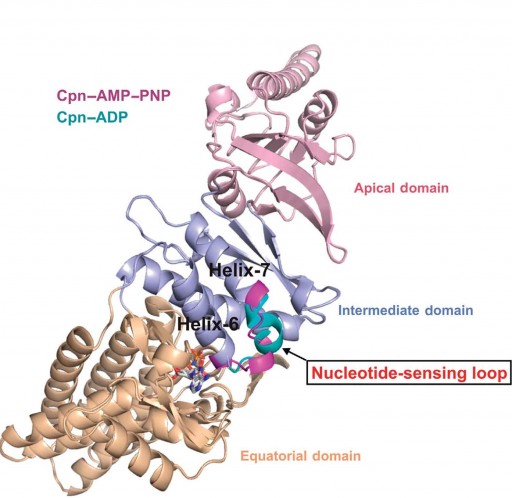
Berkeley Lab researchers at the Advanced Light Source have discovered a nucleotide-sensing loop that synchronizes conformational changes in the three domains of group II chaperonin for the proper folding of other proteins (credit: Berkeley Lab)
Researchers with the U.S. Department of Energy (DOE)’s Lawrence Berkeley National Laboratory (Berkeley Lab) have determined the crystal structure of a critical control element within chaperonin, the protein complex responsible for the correct folding of other proteins, using the exceptionally bright and powerful x-ray beams of the Advanced Light Source.
Proteins are able to fold themselves into a dazzling multitude of shapes and forms that enable them to carry out an equally dazzling multitude of functions fundamental to life. Incorrect or “misfolding” of proteins has been linked to many diseases, including Alzheimer’s, Parkinson’s and some forms of cancer.
“We identified, for the first time, a region within group II chaperonins we call the nucleotide-sensing loop, which detects the presence of the ATP molecules that fuel the chaperonin folding motion,” says Paul Adams, a bioengineer with Berkeley Lab’s Physical Biosciences Division and leading authority on x-ray crystallography, who led this work.
“We knew that ATP hydrolysis is important for promoting protein folding, but we did not know how ATP activity was sensed and communicated.”
Chaperonins promote the proper folding of newly translated proteins and proteins that have been stress-denatured — meaning they’ve lost their structure — by encapsulating them inside a protective chamber formed from two rings of molecular complexes stacked back-to-back.
“We obtained crystal structures at sufficient resolution to allow us to examine, in detail, the effects that changes in nucleotides states have on ATP binding and hydrolysis in group II chaperonins,” Adams says. “From these structures we see that the nucleotide-sensing loop monitors ATP binding sites for changes and communicates this information throughout the chaperonin.
“Functional analysis further suggests that the nucleotide-sensing loop region uses this information to control the rate of ATP binding and hydrolysis, which in turn controls the timing of the protein folding reaction.”
“The strong relationship between incorrectly folded proteins and pathological states is well documented,” Adams says. “Since ATP hydrolysis is required for protein folding, it could be possible to engineer a nucleotide-sensing loop that promotes slower or faster protein folding activity in a given chaperonin. This could, for example, be used to increase the protein folding activity of human chaperonin, or perhaps reduce the cellular accumulation of misfolded proteins that can cause disease and other problems.”
Ref.: Jose H Pereira, et al., Mechanism of nucleotide sensing in group II chaperonins, The EMBO Journal, 2012; [DOI:10.1038/emboj.2011.468]